Combined Effects of Compost and Medicago Sativa in Recovery a PCB Contaminated Soil
Abstract
:1. Introduction
2. Materials and Methods
2.1. Soil Collection and Compost Description
2.2. Microcosm Set up
- -
- Historically contaminated soil = HCS;
- -
- Historically contaminated soil + Alfalfa = Plant;
- -
- Historically contaminated soil + Compost = Compost;
- -
- Historically contaminated soil + Compost + Alfalfa = Compost +Plant
2.3. Chemical Analysis of Specific PCB Congeners
2.4. Microbiological Analyses
2.5. Structure of Microbial Community Assessed by ELFAs
2.6. Microbial Community Structure Assessed by FISH
2.7. Statistical Analysis
3. Results
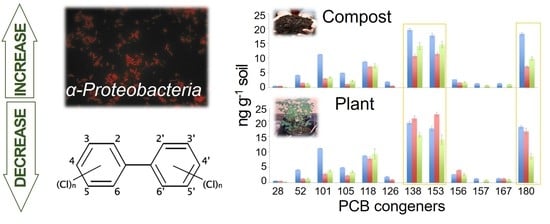
3.1. Chemical Analysis
3.2. Microbial Abundance, Cell Viability and Dehydrogenase Activity
3.3. Microbial Community Composition Evaluated Using ELFAs
3.4. Microbial Community Structure Analysed Using FISH
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Laudicina, V.A.; Dennis, P.G.; Palazzolo, E.; Badalucco, L. Key biochemical attributes to assess soil ecosystem sustainability. In Environmental Protection Strategies for Sustainable Development; Malik, A., Grohmann, E., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 193–227. [Google Scholar]
- Barra Caracciolo, A.; Bottoni, P.; Grenni, P. Microcosm studies to evaluate microbial potential to degrade pollutants in soil and water ecosystems. Microchem. J. 2013, 107, 126–130. [Google Scholar] [CrossRef]
- Babut, M.; Arts, G.H.; Barra Caracciolo, A.; Domange, N.; Friberg, N.; Gouy, V.; Grung, M.; Lagadic, L.; Martin-Laurent, F.; Mazzella, N.; et al. Pesticide risk assessment and management in a globally changing world – Report from a European interdisciplinary Workshop. Environ. Sci. Pollut. Res. 2013, 20, 8298–8312. [Google Scholar] [CrossRef] [Green Version]
- Tehrani, R.; Van Aken, B. Hydroxylated polychlorinated biphenyls in the environment: Sources, fate, and toxicities. Environ. Sci. Pollut. Res. 2014, 21, 6334–6345. [Google Scholar] [CrossRef]
- Gomes, H.I.; Dias-Ferreira, C.; Ribeiro, A.B. Overview of in situ and ex situ remediation technologies for PCB-contaminated soils and sediments and obstacles for full-scale application. Sci. Total. Environ. 2013, 445–446, 237–260. [Google Scholar] [CrossRef]
- U.S. EPA. Superfund Green Remediation Strategy, U.S. Environmental Protection Agency, Office of Solid Waste and Emergency Response, Office of Superfund Remediation and Technology Innovation. 2010. Available online: https://www.epa.gov/sites/production/files/2016-01/documents/175857.pdf (accessed on 10 January 2019).
- Song, M.; Luoa, C.; Li, F.; Jiang, L.; Wang, Y.; Zhangd, D. Anaerobic degradation of Polychlorinated Biphenyls (PCBs) and Polychlorinated Biphenyls Ethers (PBDEs), and microbial community dynamics of electronic waste-contaminated soil. Sci. Total. Environ. 2015, 502, 426–433. [Google Scholar] [CrossRef]
- Semple, K.T.; Reid, B.J.; Fermor, T.R. Impact of composting strategies on the treatment of soils contaminated with organic pollutants. Environ. Pollut. 2001, 112, 269–283. [Google Scholar] [CrossRef]
- Wenzel, W.W. Rhizosphere processes and management in plant-assisted bioremediation (phytoremediation) of soils. Plant Soil 2009, 321, 385–408. [Google Scholar] [CrossRef]
- Ancona, V.; Barra Caracciolo, A.; Grenni, P.; Di Lenola, M.; Campanale, C.; Calabrese, A.; Uricchio, V.F.; Mascolo, G.; Massacci, A. Plant-assisted bioremediation of a historically PCB and heavy metal-contaminated area in Southern Italy. New Biotechnol. Part B 2017, 38, 65–73. [Google Scholar] [CrossRef]
- Ancona, V.; Barra Caracciolo, A.; Campanale, C.; De Caprariis, B.; Grenni, P.; Uricchio, V.F.; Borello, D. Gasification treatment of poplar biomass produced in a contaminated area restored using plant assisted bioremediation. J. Environ. Manag. 2019, 239, 137–141. [Google Scholar] [CrossRef]
- Fiorentino, N.; Mori, M.; Cenvinzo, V.; Duri, L.G.; Gioia, L.; Visconti, D.; Fagnano, M. Assisted phytoremediation for restoring soil fertility in contaminated and degraded land. Ital. J. Agron. 2018, 13 (Suppl. S1), 34–44. [Google Scholar]
- Pino, N.J.; Munera, M.L.; Penuela, G.A. Phytoremediation of soil contaminated with PCBs using different plants and their associated microbial communities. Int. J. Phytoremediat. 2019, 21, 316–324. [Google Scholar] [CrossRef]
- Glick, B.R. Using soil bacteria to facilitate phytoremediation. Biotechnol. Adv. 2010, 28, 367–374. [Google Scholar] [CrossRef]
- Toussaint, J.P.; Pham, T.T.; Barriault, D.; Sylvestre, M. Plant exudates promote PCB degradation by a Rhodococcal rhizobacteria. Appl. Microbiol. Biotechnol. 2012, 95, 1589–1603. [Google Scholar] [CrossRef]
- Qin, H.; Brookes, P.C.; Xu, J. Cucurbita spp. and Cucumis sativus enhance the dissipation of polychlorinated biphenyl congeners by stimulating soil microbial community development. Environ. Pollut. 2014, 184, 306–312. [Google Scholar] [CrossRef]
- Doty, S.L. Enhancing phytoremediation through the use of transgenics and endophytes. New Phytol. 2008, 179, 318–333. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Ann. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef] [Green Version]
- Dzantor, E.K.; Chekol, T.; Vough, L.R. Feasibility of using forage grasses and legumes for phytoremediation of organic pollutants. J. Environ. Sci. Health 2000, 35, 1645–1661. [Google Scholar] [CrossRef]
- Mackova, M.; Prouzova, P.; Stursa, P.; Ryslava, E.; Uhlik, O.; Beranova, K.; Rezek, J.; Kurzawova, V.; Demnerova, K.; Macek, T. Phyto/rhizoremediation studies using long-term PCB-contaminated soil. Environ. Sci. Pollut. Res. 2009, 16, 817–829. [Google Scholar] [CrossRef]
- Ding, N.; Hayat, T.; Wang, J.E.; Wang, H.Z.; Liu, X.M.; Xu, J.M. Responses of microbial community in rhizosphere soils when ryegrass was subjected to stress from PCBs. J. Soils Sediments 2011, 11, 1355–1362. [Google Scholar] [CrossRef]
- Terzaghi, E.; Zanardini, E.; Morosini, C.; Raspa, G.; Borin, S.; Mapelli, F.; Vergani, L.; Di Guardo, A. Rhizoremediation half-lives of PCBs: Role of congener composition, organic carbon forms, bioavailability, microbial activity, plant species and soil conditions, on the prediction of fate and persistence in soil. Sci. Total. Environ. 2018, 612, 544–560. [Google Scholar] [CrossRef]
- Nogues, I.; Grenni, P.; Di Lenola, M.; Passatore, L.; Guerriero, E.; Benedetti, P.; Massacci, A.; Rauseo, J.; Barra Caracciolo, A. Microcosm experiment to assess the capacity of a poplar clone to grow in a PCB-contaminated soil. Water 2019, 11, 2220. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Teng, Y.; Li, Z.G. Enhanced removal of polychlorinated biphenyls from Alfalfa rhizosphere soil in a field study: The impact of a rhizobial inoculum. Sci. Total. Environ. 2010, 408, 1007–1013. [Google Scholar] [CrossRef]
- Teng, Y.; Shen, Y.; Luo, Y.; Sun, X.; Sun, M.; Fu, D.; Li, Z.; Christie, P.J. Influence of Rhizobium meliloti on phytoremediation of polycyclic aromatic hydrocarbons by Alfalfa in an aged contaminated soil. J. Hazard. Mater. 2011, 186, 1271–1276. [Google Scholar] [CrossRef]
- Li, Y.; Liang, F.; Zhu, Y.; Wang, F. Phytoremediation of a PCB-contaminated soil by alfalfa and tall fescue single and mixed plants cultivation. J. Soils Sediments 2013, 13, 925–931. [Google Scholar] [CrossRef]
- Tu, C.; Ma, L.; Guo, P.; Song, F.; Teng, Y.; Zhang, H.; Luo, Y. Rhizoremediation of a dioxin-like PCB polluted soil by alfalfa: Dynamic characterization at temporal and spatial scale. Chemosphere 2017, 189, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Sylvestre, M.; Toussaint, J.P. Engineering microbial enzymes and plants to promote PCB degradation in soil: Current State of Knowledge. In Microbial Bioremediation of Nonmetals—Current Research; Koukkou, A.I., Ed.; Caister Academic: Norfolk, UK, 2011; pp. 177–196. [Google Scholar]
- Di Lenola, M.; Barra Caracciolo, A.; Grenni, P.; Ancona, V.; Rauseo, J.; Laudicina, V.A.; Uricchio, V.F.; Massacci, A. Effects of Apirolio addition and Alfalfa and compost treatments on the natural microbial community of a historically PCB-contaminated soil. Water Air Soil Pollut. 2018, 229, 143. [Google Scholar] [CrossRef]
- Ballschmiter, K.; Zell, M. Analysis of polychlorinated biphenyls (PCB) by glass capillary gas chromatography. Fresenius’ Z. Anal. Chem. 1980, 302, 20–31. [Google Scholar] [CrossRef]
- Whitfield Åslunda, M.L.; Rutter, A.; Reiner, K.J.; Zeeb, B.A. The effects of repeated planting, planting density, and specific transfer pathways on PCB uptake by Cucurbita pepo grown in field conditions. Sci. Total. Environ. 2008, 405, 14–25. [Google Scholar] [CrossRef]
- Barra Caracciolo, A.; Giuliano, G.; Grenni, P.; Cremisini, C.; Ciccoli, R.; Ubaldi, C. Effect of urea on degradation of terbuthylazine in soil. Environ. Toxicol. Chem. 2005, 24, 1035–1040. [Google Scholar] [CrossRef]
- Grenni, P.; Barra Caracciolo, A.; Rodríguez-Cruz, M.S.; Sanchez-Martin, M.J. Changes in the microbial activity in a soil amended with oak and pine residues and treated with linuron herbicide. Appl. Soil Ecol. 2009, 41, 2–7. [Google Scholar] [CrossRef]
- Grenni, P.; Rodríguez-Cruz, M.S.; Herrero-Hernández, E.; Marín-Benito, J.M.; Sánchez-Martín, M.J.; Barra Caracciolo, A. Effects of wood amendments on the degradation of terbuthylazine and on soil microbial community activity in a clay loam soil. Water Soil Air Pollut. 2012, 223, 5401–5412. [Google Scholar] [CrossRef] [Green Version]
- Hinojosa, M.B.; Parra, A.; Laudicina, V.A.; Moreno, J.M. Post-fire soil functionality and microbial community structure in a Mediterranean shrubland subjected to experimental drought. Sci. Total. Environ. 2014, 573, 1178–1189. [Google Scholar] [CrossRef]
- Bossio, D.A.; Scow, K.M. Impacts of carbon and flooding on soil microbial communities: Phospholipid fatty acid profiles and substrate utilization patterns. Microb. Ecol. 1998, 35, 265–278. [Google Scholar] [CrossRef]
- Zelles, L. Fatty acid patterns of phospholipids and lipopolysaccharides in the characterization of microbial communities in soil: A review. Biol. Fertil. Soils 1999, 29, 111–129. [Google Scholar] [CrossRef]
- Schutter, M.E.; Dick, R.P. Comparison of fatty acid methyl ester (FAME) methods for characterizing microbial communities. Soil. Sci. Soc. Am. J. 2000, 64, 1659–1668. [Google Scholar] [CrossRef]
- Barra Caracciolo, A.; Bustamante, M.A.; Nogues, I.; Di Lenola, M.; Luprano, M.L.; Grenni, P. Changes in microbial community structure and functioning of a semiarid soil due to the use of anaerobic digestate derived composts and rosemary plants. Geoderma 2015, 245–246, 89–97. [Google Scholar] [CrossRef]
- Di Lenola, M.; Grenni, P.; Proença, D.N.; Morais, P.V.; Barra Caracciolo, A. Comparison of two molecular methods to assess soil microbial diversity. In Soil Biological Communities and Ecosystem Resilience; Lukac, M., Grenni, P., Gamboni, M., Eds.; Springer International Edition: Berlin, Germany, 2017; pp. 25–42. [Google Scholar]
- Barra Caracciolo, A.; Grenni, P.; Cupo, C.; Rossetti, S. In situ analysis of native microbial communities in complex samples with high particulate loads. FEMS Microbiol. Lett. 2005, 253, 55–58. [Google Scholar] [CrossRef] [Green Version]
- Greuter, D.; Loy, A.; Horn, M.; Rattei, T. probeBase-an online resource for rRNA-targeted oligonucleotide probes and primers: New features 2016. Nucleic Acids Res. 2016, 44, D586–D589. [Google Scholar] [CrossRef]
- Amann, R.; Fuchs, B.M. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 2008, 6, 339–348. [Google Scholar] [CrossRef]
- Doick, K.J.; Klingelmann, E.; Buranuel, P.; Jones, K.C.; Semple, K.T. Long-term fate of polychlorinated biphenyls and polycyclic aromatic hydrocarbons in an agricultural soil. Environ. Sci. Technol. 2005, 39, 3663–3670. [Google Scholar] [CrossRef]
- Chaudhry, Q.; Blom-Zandstra, M.; Gupta, S.K.; Joner, E.J. Utilising the synergy between plants and rhizosphere microorganisms to enhance breakdown of organic pollutants in the environment. Environ. Sci. Pollut. Res. Int. 2005, 12, 34–48. [Google Scholar] [CrossRef]
- Field, J.A.; Sierra-Alvarez, R. Microbial transformation and degradation of polychlorinated biphenyls. Environ. Pollut. 2008, 155, 1–12. [Google Scholar] [CrossRef]
- Meggo, R.E.; Schnoor, J.L. Cleaning Polychlorinated Biphenyl (PCB) Contaminated Garden Soil by Phytoremediation. Environ. Sci. 2013, 1, 33–52. [Google Scholar] [CrossRef] [Green Version]
- White, J.C.; Kottler, B.D. Citrate-mediated increase in the uptake of weathered 2,2-bis(p-chlorophenyl) 1,1-dichloroethylene residues by plants. Environ. Toxicol. Chem. 2002, 21, 550–556. [Google Scholar] [CrossRef]
- White, J.C.; Mattina, M.I.; Lee, W.-Y.; Eitzer, B.D.; Mattina, M.I. Role of organic acids in enhancing the desorption and uptake of weathered p,p′-DDE by Cucurbita pepo. Environ. Pollut. 2003, 124, 71–80. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, S.Z.; Shan, X.Q.; Zhu, Y.G. Oxalate and root exudates enhance the desorption of p,pV-DDT from soils. Chemosphere 2006, 63, 1273–1279. [Google Scholar] [CrossRef]
- Ros, M.; Hernández, M.T.; García, C. Soil microbial activity after restoration of a semiarid soil by organic amendments. Soil Biol. Biochem. 2003, 35, 463–469. [Google Scholar] [CrossRef]
- Tejada, M.; Hernández, M.T.; García, C. Application of two organic amendments on soil restoration: Effects on the soil biological properties. J. Environ. Qual. 2006, 35, 1010–1017. [Google Scholar] [CrossRef]
- Bastida, F.; Zsolnay, A.; Hernández, T.; García, C. Past, present and future of soil quality indices: A biological perspective. Geoderma 2008, 147, 159–171. [Google Scholar] [CrossRef]
- Magee, K.D.; Michael, A.; Ullah, H.; Dutta, S.K. Dechlorination of PCB in the presence of plan nitrate reductase. Environ. Toxicol. Pharm. 2008, 25, 144–147. [Google Scholar] [CrossRef]
- Macek, T.; Mackova, M.; Kas, J. Exploitation of plants for the removal of organics in environmental remediation. Biotechnol. Adv. 2000, 18, 23–34. [Google Scholar] [CrossRef]
- Leigh, M.B.; Prouzová, P.; Macková, M.; MAcek, T.; Nagle, D.P.; Fletcher, J.S. Polychlorinated biphenyl (PCB)-degrading bacteria associated with trees in a PCB-contaminated site. Appl. Environ. Microbiol. 2006, 72, 2331–2342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuiper, I.; Lagendijk, E.L.; Bloemberg, G.V.; Lugtenberg, B.J. Rhizoremediation: A beneficial plant-microbe interaction. Mol. Plant-Microbe Interact. 2004, 17, 6–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yateem, A.; Al-Sharrah, T.; Bin-Haji, A. Investigation of microbes in the rhizosphere of selected grasses for rhizoremediation of hydrocarbon-contaminated soils. Soil Sediment Contam. 2007, 16, 269–280. [Google Scholar] [CrossRef]
- van Elsas, J.D.; Bailey, M.J. The ecology of transfer of mobile genetic elements. FEMS Microbiol. Ecol. 2002, 42, 187–197. [Google Scholar] [CrossRef]
- Huelster, A.; Mueller, J.F.; Marschner, H. Soil-plant transfer of polychlorinated dibenzo-p-dioxins and dibenzofurans to vegetables of the cucumber family (Cucurbitaceae). Environ. Sci. Technol. 1994, 28, 1110–1115. [Google Scholar] [CrossRef]
- Ficko, S.A.; Rutter, A.; Zeeb, B.A. Potential for phytoextraction of PCBs from contaminated soils using weeds. Sci. Total. Environ. 2010, 408, 3469–3476. [Google Scholar] [CrossRef]
- Crecchio, C.; Curci, M.; Pizzigallo, M.D.R.; Ricciuti, P.; Ruggiero, P. Effects of municipal solid waste compost amendments on soil enzyme activities and bacterial genetic diversity. Soil Biol. Biochem. 2004, 36, 1595–1605. [Google Scholar] [CrossRef]
- Stella, T.; Covino, S.; Burianová, E.; Filipová, A.; Křesinová, Z.; Voříšková, J. Chemical and microbiological characterization of an aged PCB-contaminated soil. Sci. Total. Environ. 2015, 533, 177–186. [Google Scholar] [CrossRef]
- Thijs, S.; Sillen, W.; Rineau, F.; Weyens, N.; Vangronsveld, J. Towards an Enhanced Understanding of Plant–Microbiome Interactions to Improve Phytoremediation: Engineering the Metaorganism. Front. Microbiol. 2016, 7, 341. [Google Scholar] [CrossRef]
- Gallego-Giraldo, L.; Jikumaru, Y.; Kamiya, Y.; Tang, Y.; Dixon, R.A. Selective lignin downregulation leads to constitutive defense response expression in Alfalfa (Medicago sativa L.). New Phytol. 2011, 190, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Pinkart, H.C.; Ringelberg, D.B.; Piceno, Y.M.; Macnaughton, S.J.; White, D.C. Biochemical approaches to biomass measurements and community structure analysis. In Manual of Environmental Microbiology; Hurst, C.J., Crawford, R.L., Knudsen, G.R., McInerney, M.J., Stetzenbach, L.D., Eds.; American Society for Microbiology Press: Washington, DC, USA, 2002; pp. 101–113. [Google Scholar]
- Ben-David, E.A.; Zaady, E.; Sher, Y.; Nejidat, A. Assessment of the spatial distribution of soil microbial communities in patchy arid and semi-arid landscapes of the Negev Desert using combined PLFA and DGGE analyses. FEMS Microbiol. Ecol. 2011, 76, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Nannipieri, P.; Ascher, J.; Ceccherini, M.T.; Landi, L.; Pietramellara, G.; Renella, G. Microbial diversity and soil functions. Eur. J. Soil Sci. 2013, 54, 655–670. [Google Scholar] [CrossRef]
- Frostegård, Å.; Tunlid, A.; Bååth, E. Use and misuse of PLFA measurements in soils. Soil. Biol. Biochem. 2011, 43, 1621–1625. [Google Scholar] [CrossRef]
- Godoi, I.; Sene, L.; Barra Caracciolo, A. Assessment of the bacterial community structure in a Brazilian clay soil treated with atrazine. Ann. Microbiol. 2014, 64, 307–311. [Google Scholar] [CrossRef]
- Yao, F.; Yang, S.; Wang, Z.; Wang, X.; Ye, J.; Wang, X.; De Bruyn, J.M.; Feng, X.; Jiang, Y.; Li, H. Microbial taxa distribution is associated with ecological trophic cascades along an elevation gradient. Front. Microbiol. 2017, 8, 2071. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.K.; Munro, S.; Potts, J.M.; Millard, P. Influence of grass species and soil type on rhizosphere microbial community structure in grassland soils. Appl. Soil. Ecol. 2007, 36, 147–155. [Google Scholar] [CrossRef]
- Buée, M.; De Boer, W.; Martin, F.; van Overbeek, L.; Jurkevitch, E. The rhizosphere zoo: An overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 2009, 321, 189–212. [Google Scholar] [CrossRef]
- Bedard, D.L.; Wagner, R.E.; Brennan, M.J.; Haberl, M.L.; Brown, J.F. Extensive degradation of Aroclors and environmentally transformed polychlorinated biphenyls by Alcaligenes eutrophus H850. Appl. Environ. Microbiol. 1987, 53, 1094–1102. [Google Scholar] [CrossRef] [Green Version]
- Komancova, M.; Jurcova, I.; Kochankova, L.; Burkhard, J. Metabolic pathways of polychlorinated biphenyls degradation by Pseudomonas spp. Chemosphere 2003, 50, 537–543. [Google Scholar] [CrossRef]







| Italian Legal Limits | ||
|---|---|---|
| E. coli (CFU g−1) | <25 | ≤1000 |
| Salmonella (CFU g−1) | Absent | Absent |
| Microbial abundance (No. cells g−1) | 1.0 × 1010 | - |
| Cell viability (% live cells/live + dead) | 3% | - |
| Dehydrogenase activity (μg TPF g−1) | 550 | - |
| Probe Name | Short Name | Specificity * | Sequence from 5′ to 3′ | Target Molecule and rRNA Position | Stringency (%) |
|---|---|---|---|---|---|
| ARCH915 | ARCH | Archaea | GTGCTCCCCCGCCAATTCCT | 16S rRNA, 915-934 | 20 |
| EUB338 ** (EUB) | EUB | Most Bacteria | GCTGCCTCCCGTAGGAGT | 16S rRNA, 338-355 | 20 |
| EUB338 II ** | EUB | Planctomycetales | GCAGCCACCCGTAGGTGT | 16S rRNA, 338-355 | 20 |
| EUB338 III ** | EUB | Verrucomicrobiales | GCT GCC ACC CGT AGG TGT | 16S rRNA, 338-355 | 20 |
| ALF1b | A | α -Proteobacteria, some Deltaproteobacteria, Spirochaetes | CGT TCG (CT) TC TGA GCC AG | 16S rRNA, 19-35 | 20 |
| BET42a § | Β | β-Proteobacteria | GCC TTC CCA CTT CGT TT | 23S rRNA, 1027-1043 | 35 |
| GAM42a ° | Γ | γ-Proteobacteria | GCC TTC CCA CAT CGT TT | 23S rRNA, 1027-1043 | 35 |
| DELTA495a ^ | Δ | Most δ-Proteobacteria, most Gemmatimonadetes | AGT TAG CCG GTG CTT CCT | 16S rRNA, 495-512 | 35 |
| DELTA495b ^ | Some δ-Proteobacteria | AGT TAG CCG GCG CTT CCT | 16S rRNA, 495-512 | 35 | |
| DELTA495c ^ | Some δ-Proteobacteria | AAT TAG CCG GTG CTT CCT | 16S rRNA, 495-512 | 35 | |
| CF319a | CF | Most Flavobacteria, some Bacteroidetes, some Sphingobacteria | TGG TCC GTG TCT CAG TAC | 16S rRNA, 319-336 | 35 |
| HGC69A | HGC | Actinobacteria (Gram-positive bacteria with high DNA G+C content) | TAT AGT TAC CAC CGC CGT | 23S rRNA, 1901-1918 | 35 |
| LGC354A ** | LGC | Firmicutes (Gram-positive bacteria with low G+C content) | TGG AAG ATT CCC TAC TGC | 16S rRNA, 354-371 | 35 |
| LGC354B ** | CGG AAG ATT CCC TAC TGC | 16S rRNA, 354-371 | 35 | ||
| LGC354C ** | CCG AAG ATT CCC TAC TGC | 16S rRNA, 354-371 | 35 |
| N. Chlorine | tri-Cl | tetra-Cl | penta-Cl | hexa-Cl | hepta-Cl |
|---|---|---|---|---|---|
| HCS | 66% ± 0.66 | 66% ± 0.74 | 36% ± 0.00 | 2% ± 0.12 | 29% ± 0.01 |
| Compost | 60% ± 0.39 | 69% ± 0.24 | 52% ± 2.54 | 25% ± 0.69 | 46% ± 0.48 |
| Plant | 60% ± 0.73 | 70% ± 0.15 | 37% ± 5.98 | 20% ± 0.93 | 52% ± 1.16 |
| Compost + Plant | 54% ± 0.91 | 71% ± 0.23 | 53% ± 2.20 | 16% ± 1.32 | 44% ± 1.44 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Lenola, M.; Barra Caracciolo, A.; Ancona, V.; Laudicina, V.A.; Garbini, G.L.; Mascolo, G.; Grenni, P. Combined Effects of Compost and Medicago Sativa in Recovery a PCB Contaminated Soil. Water 2020, 12, 860. https://doi.org/10.3390/w12030860
Di Lenola M, Barra Caracciolo A, Ancona V, Laudicina VA, Garbini GL, Mascolo G, Grenni P. Combined Effects of Compost and Medicago Sativa in Recovery a PCB Contaminated Soil. Water. 2020; 12(3):860. https://doi.org/10.3390/w12030860
Chicago/Turabian StyleDi Lenola, Martina, Anna Barra Caracciolo, Valeria Ancona, Vito Armando Laudicina, Gian Luigi Garbini, Giuseppe Mascolo, and Paola Grenni. 2020. "Combined Effects of Compost and Medicago Sativa in Recovery a PCB Contaminated Soil" Water 12, no. 3: 860. https://doi.org/10.3390/w12030860