Phylogenomics and systematics of botryllid ascidians, and implications for the evolution of allorecognition
- 1Life Sciences Concentration, Soka University of America, Aliso Viejo, CA, United States
- 2Department of Scientific Computing, Florida State University, Tallahassee, FL, United States
- 3Department of Biological Science, Florida State University, Tallahassee, FL, United States
- 4Biology Department and Estuary and Ocean Science Center, San Francisco State University, Tiburon, CA, United States
- 5Posgrado en Ciencias del Mar y Limnología, Universidad Nacional Autónoma de México, Ciudad de México, Mexico
- 6Unidad Multidisciplinaria de Docencia e Investigación Sisal (UMDI-Sisal), Facultad de Ciencias Nacional Autónoma de México, Ciudad de México, Mexico
- 7Department of Biosciences, Biotechnologies and Environment, University of Bari “Aldo Moro”, Bari, Italy
- 8Institute of Biomembranes, Bioenergetics and Molecular Biotechnologies (IBIOM), Consiglio Nazionale delle Ricerche, Bari, Italy
- 9Consorzio Nazionale Interuniversitario per le Scienze del Mare (CoNISMa), Roma, Italy
Allorecognition, the ability of an organism to distinguish kin from non-kin, or self from non-self, has been studied extensively in a group of invertebrate chordates, the colonial ascidians called botryllids (Subphylum Tunicata, Class Ascidiacea, Family Styelidae). When two conspecific botryllid colonies come in contact, there are two potential outcomes to an allorecognition reaction: fusion or rejection. The rejection outcome of allorecognition varies by species, and has been classified by type (referred to as R-Type). R-Type is defined according to how far the fusion process progresses before the rejection begins, since the rejection reaction appears as an interference of the fusion process. Here, we map the evolution of R-Types onto an extended and robust phylogeny of the botryllids. In this study, we have reconstructed the largest phylogenomic tree of botryllids, including 97 samples and more than 40 different species, and mapped on it nine of the 13 species for which the R-Type is known. Based on the R-Type known in a single outgroup species (Symplegma reptans), we infer that at least R-Type B and E-like could be ancestral to the Botrylloides/Botryllus group. However, the application of ancestral character state reconstructions does not provide conclusive results since several clades show more than one equiparsimonious R-Type state. Notably, all R-Type A species are clustered together and certainly evolved later than other R-Types. Our phylogenomic tree has been built on 177 nuclear loci and nearly all clades are well supported. Moreover, our phylogenetic analyses also take into account the results of species delimitation analyses based on the mitochondrial COI gene and of careful morphological analyses of the samples. The implementation of this integrated taxonomic approach, combining morphological as well as nuclear and mitochondrial data, has allowed the description of six new species, and the identification of a number of putative unnamed taxa. Thus, our results also demonstrate the existence of an unexplored hidden diversity within botryllids.
Introduction
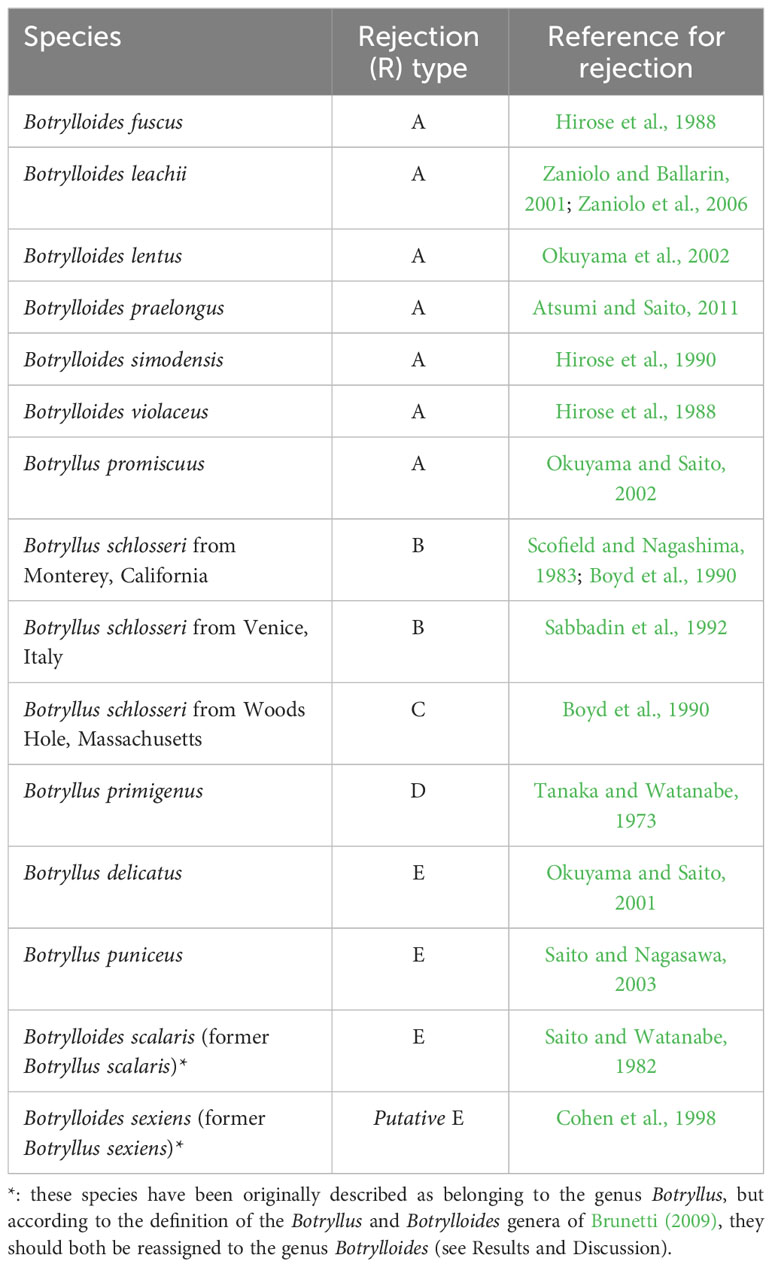
Allorecognition is the ability of an organism to distinguish kin from non-kin, or self from non-self and is found in all major groups of organisms, from eubacteria to plants (Buss, 1982). It has been studied extensively in a focal group of invertebrate chordates, the colonial ascidians called botryllids (Subphylum Tunicata, Class Ascidiacea, Family Styelidae), also widely used as models in immunobiology, angiogenesis, asexual reproduction and whole body regeneration (Lauzon et al., 2013; Voskoboynik and Weissman, 2015; Blanchoud et al., 2018; Ballarin et al., 2021; Nourizadeh et al., 2021). When two conspecific botryllid colonies come in contact, there are two potential outcomes to an allorecognition reaction: fusion or rejection (the overall phenomenon is called “colony specificity”). In fusion, colonies that share alleles at allorecognition loci physically merge (Saito et al., 1994). In rejection, colonies that do not share these allorecognition alleles undergo cytotoxic reactions and erect borders to prevent further interaction (Saito et al., 1994). In botryllid colonies, zooids (single animals genetically identical to all other zooids in the colony) are covered with a common tunic and are connected to the other zooids by a ramified vascular network (De Tomaso et al., 2005). At the colony edge, the terminals of the vascular vessels form several vascular ampullae (De Tomaso et al., 2005). Fusion and rejection processes mainly involve the ampullae and the tunic covering them, including blood within the ampullae (De Tomaso et al., 2005). Across botryllids, the fusion process is very similar and occurs through five stages, but the rejection processes vary between species (Saito et al., 1994 and references therein). According to Figure 7 of Saito et al. (1994), five types of rejection (R-Type A, R-Type B, R-Type C, R-Type D, and R-Type E) can be recognized: they are classified by how far the fusion process progresses before the rejection begins, since the rejection reaction appears as an interference of the fusion process. A scheme depicting the five R-Types, redrawn from Figure 7 of Saito et al. (1994), is shown in Figure 1.
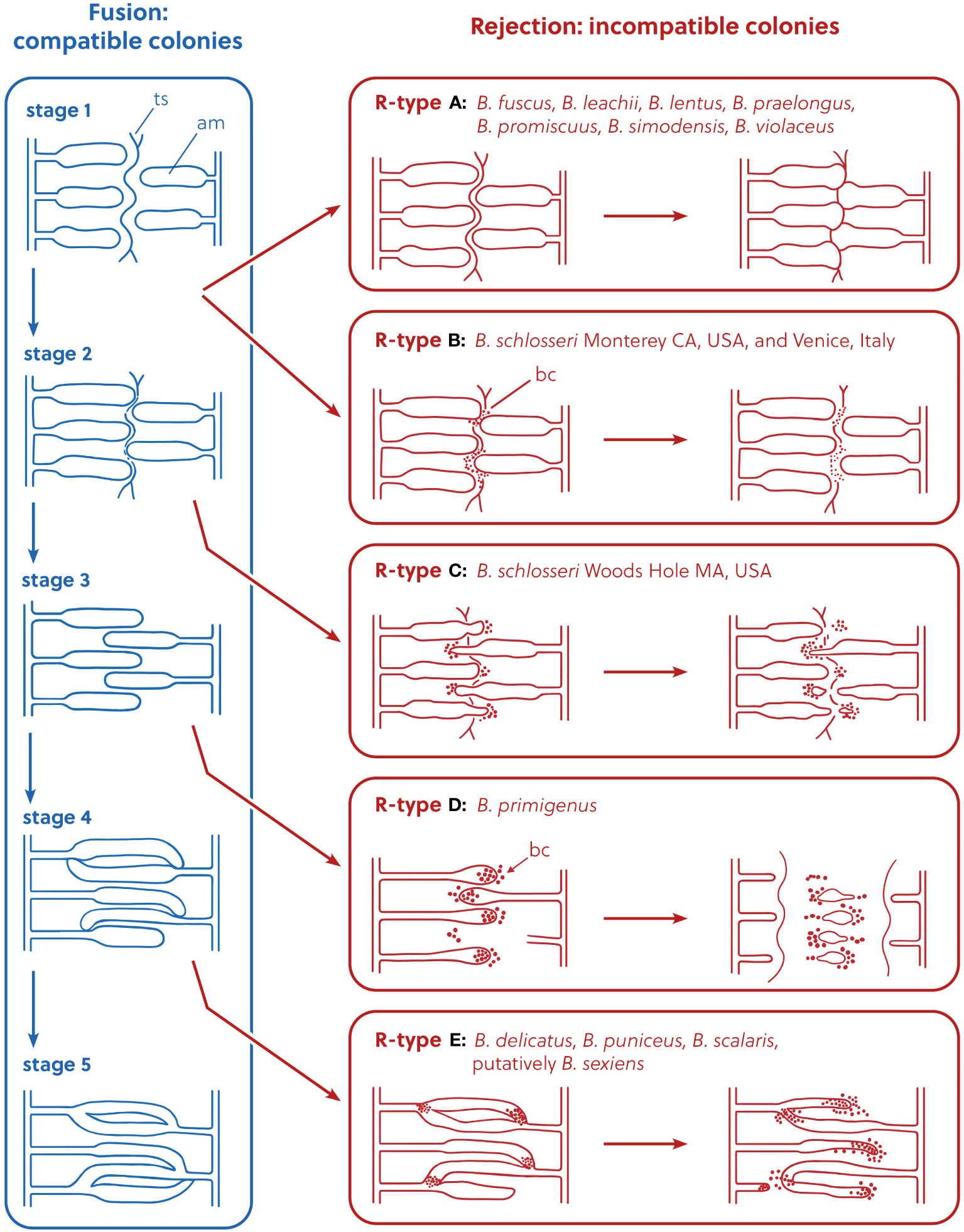
Figure 1 Representation of the allorejection process, R-Types A–E, redrawn and modified from Saito et al. (1994), Figure 7. In the rejection panels, the diagram is not intended to show the exact position of the ampullae, but to describe their relative position compared to the stage of origin in the fusion process. ts, tunic surface; am, ampulla; bc, blood cell.
R-Type A rejection occurs at the first stage of fusion (Saito et al., 1994). In this type of rejection, the outer surface of the animal’s body (the tunic) is the site of rejection: fusion of the tunics of the two colonies occurs first, and only at limited points of contact, and the ampullae do not fuse. Then, cytotoxic reactions occur within the tunic between blood cells of the opposing colonies (Saito, 2003). In the final phase of the rejection reaction, a new border composed of a thin, fibrous material is formed between the opposing colonies. R-Type B rejection also occurs at the first stage of fusion, but in its final phase the blood cells become black and necrotic, and are visible without a microscope. R-Type C rejection occurs at the second stage of fusion: as in R-Type A and B, tunic fusion only occurs in small areas around ampullar tips. After blood cells infiltrate around the tips of the penetrating ampullae, these ampullae begin to shrink and withdraw or are amputated. R-Type D rejection occurs during the third stage of fusion, and complete fusion of the outer tunic occurs first. Then, we see penetration of one colony’s ampullae into the opposing colony’s tunic (but, without fusion between opposing colonies’ ampullae). Just like in R-Types A–C, then cytotoxic reactions occur within the tunic between blood cells of the opposing colonies. When the colonies reject, a dark necrotic reaction is observed around the interacting vessels and cells. Vessels are then pinched off and abandoned in a necrotic area that is demarcated by a thick fibrous barrier between the two colonies. This barrier between colonies is not seen in R-Type C rejections. R-Type E is at the most extreme end of the behavioral spectrum of botryllid allorecognition: it consists of the complete membrane fusion of the circulatory systems of opposing colonies and subsequent exchange of blood cells between the two colonies. Cells in the blood interact between the two colonies and these interacting cells turn dark and lyse. Parts of the shared circulatory system and tunic area become necrotic and the two colonies both shrink away from the zone of interaction. Fused circulatory elements may be pinched off from the parent colony and left to degenerate in a necrotic zone (Saito et al., 1994).
The R-Type has been characterized only in some botryllids and current knowledge shows that it is species-specific (Saito et al., 1994; Hirose et al., 2002; Saito, 2003) (Table 1). As an exception to this species-specificity, Botryllus schlosseri populations from California, USA, show R-Type B rejection (Saito et al., 1994), as do populations from Venice, Italy (Sabbadin et al., 1992), whereas B. schlosseri populations from Massachusetts, USA, show R-Type C rejection (Saito et al., 1994). However, this exception is very probably only ostensible and related to the species-complex status of B. schlosseri, discovered and partially solved (Lopez-Legentil et al., 2006; Bock et al., 2012; Yund et al., 2015; Nydam et al., 2017a; Brunetti et al., 2020; Salonna et al., 2021) only after Saito’s study on the B. schlosseri rejection process. The species-specificity of the R-type suggests that the pathways, gene expression or gene regulatory mechanisms involved in the rejection process are at least partially different between the different botryllids. Therefore, these R-types should be placed in a phylogenetic context, in order to understand the evolution of allorecognition in botryllids. An 18S rDNA (Cohen et al., 1998) and a mitochondrial cytochrome oxidase COI (mtCOI) (Salonna et al., 2021) botryllid phylogeny provide a first foundation to solve this issue. In particular, Cohen et al.’s mapped the allorecognition behavioral variation on an 18S rDNA tree comprising nine botryllids, and found R-Type E species at the base of the botryllid tree, with R-Type D species evolving next, and R-Type A species evolving most recently (Figure 2A, summarizing the results of Figure 3 of Cohen et al., 1998). Thus, Cohen et al. (1998) inferred that more derived species evolved more external allorecognition responses and that the R-Type E, i.e., the most internal form of allorecognition, was the ancestral condition. Their hypothesis does not strictly follow the parsimony criterion and was not made applying a specific “ancestral state reconstruction” analysis, however Cohen et al. underlined that the identification of internal allorecognition as the ancestral state is compatible with observations on the allogeneic blood reactions in solitary ascidians. In Cohen et al.’s tree, the phylogenetic relationship between R-Type E and the other R-Types is supported with a 80% bootstrap confidence value in the Maximum Likelihood tree (Figure 2A), but only with a 60% bootstrap confidence value in the Maximum Parsimony tree (see Cohen et al., 1998, Figure 2). If we map the R-type on the Salonna’s mtCOI tree, an R-Type E species is again found to be an early diverging lineage (Figure 2B), evolving just after the node of Botrylloides conchyliatus (Salonna et al., 2021) for which there is no allorecognition information. However, the position of this R-Type E species is not well-supported. In general, most nodes of this mtCOI tree remain unresolved. Indeed, even the clade including R-Type A and R-Type D species is not well-supported (Figure 2B). Only R-Type B and C, so far characterized only in the B. schlosseri species complex (Table 1), are in a well-supported clade with no other rejection types (Figure 2B).
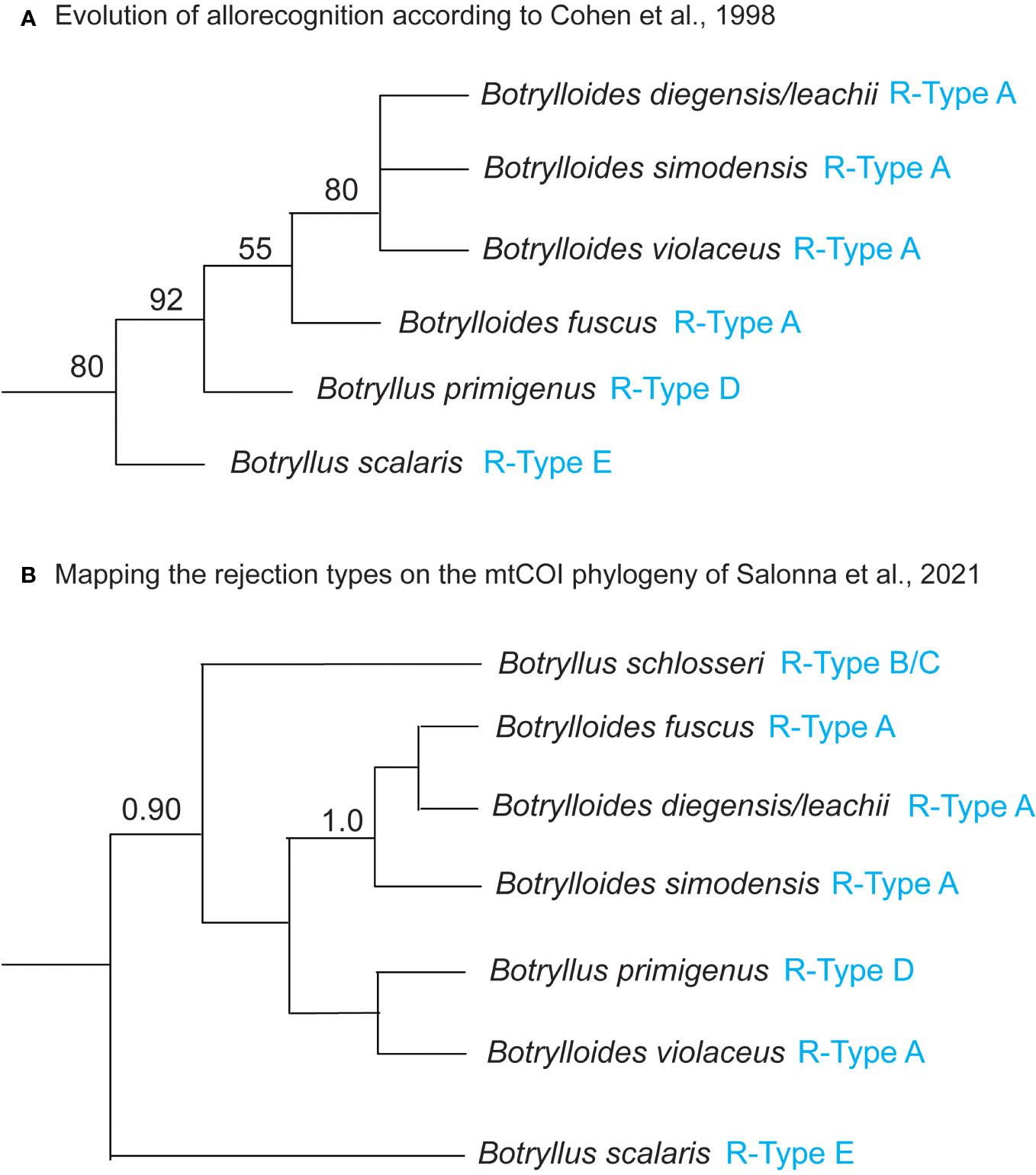
Figure 2 (A) Evolution of allorecognition in botryllids according to Cohen et al. (1998). 18S rDNA Maximum Likelihood tree with bootstrap percentage values at the nodes, reproduced from Cohen et al. (1998) Figure 3 and including only species for which the R-type is known. (B) Mapping the rejection types on the mtCOI phylogenetic tree of Salonna et al. (2021). Mitochondrial COI Bayesian majority rule consensus tree reconstructed from the “Elongated-856nt” alignment, reproduced from Salonna et al. (2021) Figure 2 and including only species for which the R-type is known. Bayesian posterior probabilities at the nodes are only represented if they are ≥ 0.90. R-types (from Table 1) are indicated in blue. Botrylloides diegensis/leachii: based on Viard et al. (2019), all published mtCOI sequences of Botrylloides leachii should actually belong to Botrylloides diegensis. Waiting for confirmation by more detailed morphological analyses, here we labeled the 18S rDNA and the mtCOI sequences as Botrylloides diegensis/leachii. See discussion in the section “Mapping mtCOI Species Delimitation on the Phylogenomic Tree”.
In this study we have reconstructed the botryllid phylogeny using 177 Anchored Hybrid Enrichment (AHE) nuclear loci selected among the 200 nuclear probes developed in Nydam et al. (2021), in order to resolve the deepest unresolved nodes present in the single-locus botryllid phylogenies based on 18S rRNA and mtCOI. The obtained phylogeny has 97 specimens and includes: 21 samples specifically collected for this study; most of the specimens analyzed in Salonna et al. (2021) and in Palomino-Alvarez et al. (2022); as well as the entire sample dataset of Nydam et al. (2021). Therefore, it represents the largest known botryllid diversity. Since our phylogeny contains nine of the 13 species that have been characterized for rejection type, it is also particularly appropriate for reconstructing the evolutionary history of the allorecognition reaction in botryllids.
Materials and methods
Sample collection
A total of 21 botryllid samples were collected for this study and analyzed together with 46 samples published in Nydam et al. (2021), 18 samples published in Salonna et al. (2021) and 12 samples published in Palomino-Alvarez et al. (2022). All these samples (total of 97) were used to build the phylogenomic trees described below and they are listed in Supplementary Table 1, together with collection information and the related mtCOI Accession numbers. Photographs and morphological details of the samples that were collected as part of the current study are in Supplementary File 1.
For the 21 new samples, a small piece of colony was removed from each sample in the field, cleaned to remove algae and other contaminants, and preserved in 95% ethanol, RNAlater (Thermo-Fisher), or a DMSO solution saturated with NaCl. For the species that were morphologically described, a fragment of the colony was also relaxed using menthol crystals and subsequently preserved in 10% formalin in salt water buffered with sodium borate.
Sample identification: DNA barcoding
For 16 of the 21 new samples, we sequenced the mtCOI gene for molecular barcoding purposes. Accession numbers for these 16 sequences are denoted in Supplementary Table 1, with an asterisk. DNA was extracted using the Nucleospin Tissue Kit (Macherey Nagel). PCR was performed using either OneTaq DNA Polymerase (New England Biolabs) or Phusion High-Fidelity DNA Polymerase (New England Biolabs). OneTaq reactions comprised the following ingredients: 25 μl total reaction volume with 16.38 μl of nuclease-free water (New England Biolabs), 5 μl of 5X buffer (New England Biolabs), 0.5 μl of 10 mM dNTPs, 0.5 μl of 10 μM of each primer, 0.12 μl of OneTaq and 2 μl of DNA template (100–500 ng). Phusion reactions were as follows: 20 μl total reaction volume with 10.8 μl of nuclease-free water (New England Biolabs), 4 μl of 5X HF buffer (New England Biolabs), 0.4 μl of 10 mM dNTPs, 0.6 μl of 100% DMSO, 1 μl of 10 μM of each primer, 0.2 μl of Phusion and 2 μl of DNA template (100–500 ng).
Each DNA sample was amplified with one of the following PCR primer pairs: LCO1490/HCO2198 (Folmer et al., 1994) or Tun_forward/Tun_reverse2 (Stefaniak et al., 2009). “Tun” primers were only used with OneTaq polymerase, following this protocol: 94°C for 1 min, 60x (94°C for 10 sec, 50°C for 30 sec, 72°C for 50 sec), 72°C for 10 min. Folmer’s primers were only used with Phusion polymerase, following this protocol: 98°C for 30 sec, 35x (98°C for 10 sec, 48°C for 30 sec, 72°C for 30 sec), 72°C for 5 min. Supplementary Table 1 reports the primer pair used for each sample first published in this study.
PCR products were purified through incubation with 1 µl each of Exonuclease I (New England Biolabs) and Antarctic Phosphatase (New England Biolabs) at 37°C for 1 hour, followed by 90°C for 10 min. All PCR products were Sanger sequenced in both directions at Eurofins Genomics (Louisville, KY) using industry standard protocols. Forward and reverse sequences were edited and combined into a consensus sequence using Codon Code Aligner (Codon Code Corporation). Consensus sequences are available on GenBank (Accession Numbers are listed in Supplementary Table 1).
Sample identification: morphological techniques
A formalin-preserved portion was saved for 10 of the 21 botryllid samples specifically collected for this study and used for morphological studies (see column “Morphology” in Supplementary Table 1). These samples were assigned to species using descriptions from the literature (Oka, 1928; Saito et al., 1981a; Saito et al., 1981b; Saito and Watanabe, 1985; Okuyama and Saito, 2001; Saito and Okuyama, 2003; Atsumi and Saito, 2011). A total of 31 morphological characters were analyzed and are summarized as follows: arrangement of systems, position of ovaries and testes, testes morphology, number of stigmatal rows, completeness of the second stigmatal row, arrangement of stigmata, shape of intestine, location of anterior edge of intestinal loop, location of anus, number of stomach folds, appearance of the stomach folds, shape of the stomach, shape and size of the pyloric caecum, number and size orders of the oral tentacles, distribution of pigment cells in the zooid, zooid length, colony color when living and after fixation, and tunic thickness (Saito and Watanabe, 1985; Saito and Okuyama, 2003; Brunetti, 2009). New described species were assigned to the genus Botryllus or Botrylloides according to the Brunetti’s definition of these genera (Brunetti, 2009).
Species delimitation
To determine where botryllid species boundaries lie, species delimitation analyses were conducted on a dataset including:
● 73 of the 77 mtCOI sequences listed in Supplementary Table 1 (the four mtCOI sequences reported with a tilde sign (~) in Supplementary Table 1 were excluded because they were too short to include in the alignment);
● 175 additional mtCOI sequences belonging primarily to the taxa represented in the phylogenetic tree but also to eight additional botryllid species and to five Symplegma species (see species with an asterisk in Supplementary Table 2 Column E).
Symplegma (Family Styelidae, Subfamily Polyzoinae) was used as an outgroup since previous morphological and molecular data have identified this genus as sister to botryllids (Berrill, 1950; Ben-Shlomo et al., 2010).
Thus, the whole analyzed mtCOI dataset comprises 248 sequences that were either generated as part of the current study or previously published in Palomino-Alvarez et al. (2022) or Salonna et al. (2021). Supplementary Table 2 reports the results of the species delimitation analyses, and includes also the GenBank Accession Numbers of the 248 analyzed sequences with the relevant publication information. A 467 bp alignment of 247 mtCOI sequences was created by the Muscle alignment algorithm in CodonCode Aligner v. 10 (CodonCode Corporation, Centerville, MA). This alignment was analyzed using two single-locus methods for species delimitation: ASAP (Puillandre et al., 2021) and bPTP (Zhang et al., 2013). ASAP (Assemble Species by Automatic Partitioning) uses pairwise genetic distances to partition putative species (named OTUs, Operational Taxonomic Unit) by identifying gaps between larger distances (presumed interspecific) and smaller distances (presumed intraspecific) (Puillandre et al., 2021). ASAP builds on the earlier program ABGD (Automatic Barcode Gap Discovery) (Puillandre et al., 2012) by removing the a priori defined maximum genetic intraspecific divergence (P) and ranking the partitions (Puillandre et al., 2021). bPTP (Bayesian Poisson Tree Process) is derived from PTP, which analyzes a user-inputed phylogenetic tree to identify regions of the tree that are transition points between branching rates consistent with a speciation model and those consistent with a coalescent model (Zhang et al., 2013). bPTP adds Bayesian support values to the putative species nodes (Zhang et al., 2013). The OTU partitions between ASAP and bPTP can differ (Ducasse et al., 2020), so we employed both methods.
We ran ASAP using all three available substitution models: p-distances, Jukes-Cantor, and Kimura 2P. The analysis was performed using ASAP web server: https://bioinfo.mnhn.fr/abi/public/asap/asapweb.html. A Maximum Likelihood (ML) tree was generated from the same 467 bp alignment of 248 sequences using RAxML HPC BlackBox (Stamatakis, 2014) on CIPRES (Cyberinfrastructure for Phylogenic Research) Science Gateway v. 3.3 (Miller et al., 2010). This ML tree was analyzed with bPTP using the web server: http://species.h-its.org/, and results from both Maximum Likelihood and Bayesian-derived partitions were obtained.
We considered a clade to be a species if at least two of the three analyses (ASAP, bPTP-ML and bPTP-Bayesian) agreed to recover that clade as a single OTU. ASAP and bPTP use different conceptual and algorithmic frameworks, and bPTP-ML and BPTP-Bayesian use different trees. We decided to use results that were supported by at least two of the three methods, as we believe that these results would be more robust than results supported by only one method.
Phylogenomic tree building
DNA extraction, library preparation, anchored hybrid enrichment (AHE), library sequencing, and raw read alignment methods were conducted on 51 samples (i.e., all samples of Supplementary Table 1 not previously published in Nydam et al., 2021) as described in Nydam et al. (2021) aside from three minor changes. All three changes are modifications of parameter values used in the automated procedure described in Hamilton et al. (2016). The changes are outlined below: first, during raw read alignment, consensus sequences were constructed from assembly clusters containing greater than an average of 37 reads (previously-used value: 250). Second, during masking, a 10-base threshold was used for masking misaligned regions (previously-used value: 14). Lastly, during trimming, 49 sequences were required to be present at a site to prevent removal of the site (previously-used value: 25). Only 177 loci were recovered after quality control and filtering. The size of these loci and the correspondence to the AHE nuclear probes and loci used in Nydam et al. (2021) is reported in Supplementary Table 3.
Phylogenomic trees were built on the whole dataset of 97 samples using two different methods. First, using RAxML v. 8.2.12 (Randomized Axelerated Maximum Likelihood) a species tree was estimated based on the sequence data from all loci concatenated together. Although concatenated, alpha-shape parameters, GTR rates, and empirical base frequencies were estimated separately for each “locus” using the -q option in RAxML; GTR Gamma was specified as the model of nucleotide substitution. Bootstrap support values for the best scoring ML tree were calculated from 100 rapid bootstrap replicates. Second, an unrooted species tree was estimated using a coalescent summary method, ASTRAL-II v.5.7.8 (Accurate Species TRee ALgorithm). Individual single-locus trees provided as input into the ASTRAL analysis were constructed in RAxML with 100 rapid bootstrap replicates using the GTR Gamma model of nucleotide substitution. The support values on the ASTRAL tree are automatically generated from quartet frequencies; the authors of ASTRAL recommend using these values instead of bootstrapping. Symplegma sp. was forced as an outgroup in both the concatenated RAxML tree and the ASTRAL tree. Additional details of these methods are available in Nydam et al. (2021).
Ancestral state reconstructions
Ancestral character state reconstructions in a parsimony framework were conducted in Mesquite v. 3.8.1 using the RAxML phylogenomic tree as described above (Maddison and Maddison, 2023). The single character “R-Type” was traced across the phylogeny, with the five possible character states being “R-Type A”, “R-Type B”, “R-Type C”, “R-Type D”, and “R-Type E”. The R-type of the outgroup Symplegma sp. can be either similar phenotypically to R-Type B or to R-Type E, although it is not known whether the molecular or mechanistic bases of these types are similar. We therefore ran three analyses: one with Symplegma sp. coded as unknown, one with Symplegma sp. coded as R-Type B, and one with Symplegma sp. coded as R-Type E. When Symplegma sp. received an “R-Type”, then 26 (26.8%) of the 97 specimens in the analysis received an “R-Type”, and 71 were classified as unknown. When Symplegma sp. did not receive an “R-Type”, then 24 (24.7%) of the 97 specimens in the analysis received an “R-Type” and 73 were classified as unknown. Botryllus sexiens is classified in the literature as “putatively” R-Type E, and in this analysis it was coded as R-Type E.
Results and discussion
Sample identification reveals a hidden botryllid diversity
A total of 21 samples were specifically collected for this study in several localities around the world, from Japan to America and Australia (Supplementary Table 1). According to an integrated taxonomy approach, their taxonomic identification was based on morphological examinations and/or mtCOI barcoding followed by species delimitation analyses. Morphological descriptions for each of these samples are in Supplementary File 2, where they are discussed together with the results of the species delimitation analyses. The overall results of species delimitation analyses are summarized with blue circles/rectangles in Figure 3 and shown in detail in Supplementary Table 2 (see also the section “Mapping Species Delimitation on the Phylogenomic Tree”).
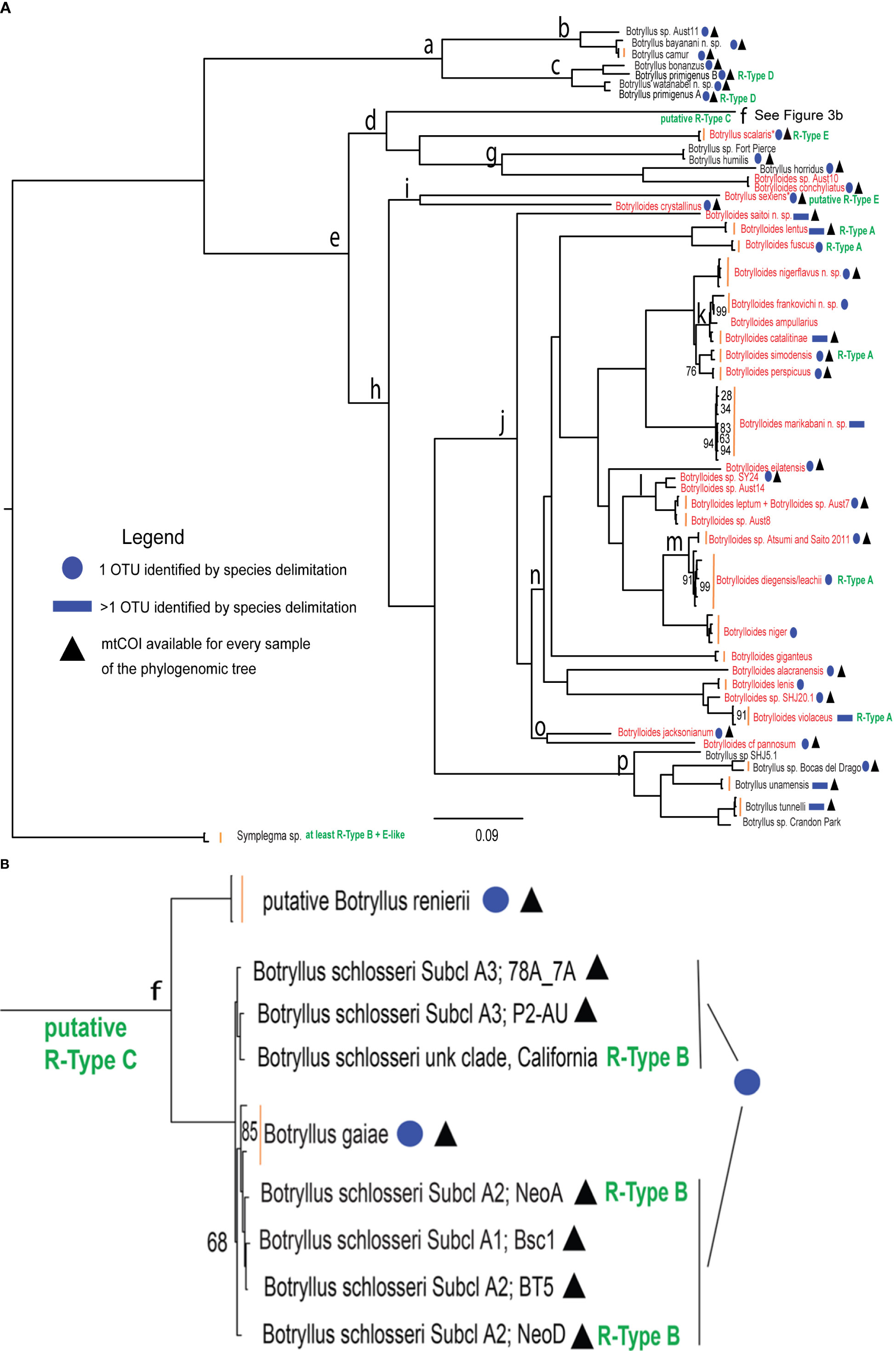
Figure 3 Phylogenomic tree of botryllid based on 177 concatenated nuclear loci and generated by RAxML. (A) Entire tree excluding clade “f”. (B) Clade “f” corresponding to the Botryllus schlosseri species complex. The scale bar indicates 0.09 substitutions per site. R-types are in green text. Botrylloides species and species proposed to be re-assigned to the genus Botrylloides are in red text. Blue circles refer to samples identified as belonging to the same OTU in the mtCOI species delimitation results shown in Supplementary Table 2. Blue rectangles refer to samples belonging to more than one OTU as identified in the mtCOI species delimitation results shown in Supplementary Table 2. Black triangles denote OTUs for which the mtCOI sequences used in species delimitation analyses are available for every sample that appears in the phylogenomic tree. Vertical orange bars group leaves of the same species. An asterisk labels Botryllus species proposed to be reassigned to the Botrylloides genus. “Subcl” indicates subclades as recognized in Botryllus schlosseri sensu Brunetti et al. (2017) by the “Elongated-856nt” mtCOI alignment analyzed in Salonna et al. (2021). Bootstrap support values are displayed at each node only when < 100%. The clade “f” branch in panel (A) was truncated at level of its internal node.
The specimens from Japan belong to five already-described species (i.e., Botrylloides lenis, Botrylloides lentus, Botrylloides simodensis, Botryllus scalaris and Botryllus sexiens) and to two new species described here for the first time: Botrylloides saitoi n. sp. and Botryllus watanabei n. sp.
No formalin-preserved fraction was available for the four new specimens from Heron Island, Australia (Botrylloides sp. Aust 7, Botrylloides sp. Aust 8, Botrylloides sp. Aust 10 and Botrylloides sp. Aust 14), and for a specimen from Florida (Botryllus sp. Crandon Park Marina, Key Biscayne, FL, USA). Moreover, a mtCOI sequence with a length suitable for species delimitation analyses was obtained only for Botrylloides sp. Aust 7 (Supplementary Tables 1 and 2). Thus, these specimens were tentatively identified based on their positions in the phylogenomic trees (Figure 3 and Supplementary File 5):
● Botrylloides sp. Aust 7 and Botrylloides sp. Aust 8 samples are in the same clade as Botrylloides leptum (inside Clade “m”) with very little nuclear genetic divergence within this clade, as shown by the very short branches. Although the mtCOI sequence of Botrylloides sp. Aust 7 is 98.3% identical to Botrylloides leptum and is included in the same OTU of Botrylloides leptum by all species delimitation analyses (blue circle in Figure 3 and Supplementary Table 2), conclusive identification will require the comparison of formalin-preserved samples of both the Heron Island species and of Botrylloides leptum.
● Botrylloides sp. Aust 10 is in the same clade as Botrylloides conchyliatus (inside Clade “g”) with very little nuclear genetic divergence between the two taxa, so this specimen was assigned to the Botrylloides genus (see section: “Phylogenomic tree and the Botryllus-only Clade “a””). Confirmation of the taxonomic assignment of this sample awaits a mtCOI barcode sequencing and morphological examination.
● Botrylloides sp. Aust 14 clusters together with Australian Botrylloides sp. SY24 (inside Clade “l”) with little nuclear genetic divergence shown. The lack of formalin-preserved samples for both samples (Supplementary Table 1; Nydam et al., 2021) and of the mtCOI sequence from Botrylloides sp. Aust 14 (Supplementary Table 1) prevents a well-defined taxonomic attribution and requires further study.
● Botryllus sp. from Crandon Park Marina (Key Biscayne, FL, USA) in a clade including only Botryllus species (inside Clade “p” of Figure 3), so it was tentatively assigned to the genus Botryllus (hereafter named “Botryllus Crandon Park”; one sample). The lack of a formalin-preserved sample and a mtCOI sequence too short for species delimitation analyses means that further study is required for a more reliable taxonomic attribution.
Two distinct new taxa were identified by species delimitation analyses as “unnamed species” since they did not group to mtCOIs associated with a given species. Moreover, they could not be assigned to an already known species, nor described as a new species, because formalin-preserved samples were not available. This is the case for Botrylloides sp. SHJ-20.1 from Shikine Island, Izu Islands, Japan (hereafter named “Botrylloides SHJ20.1”; one sample; inside Clade “n” and with a blue circle in Figure 3), and the species hereafter named “Botrylloides sp. (Atsumi and Saito, 2011)” (two samples; inside Clade “m” and with a blue circle in Figure 3). Although this last species was not formally described in Atsumi and Saito (2011), it was reported in that study as morphologically very similar to Botrylloides simodensis and genetically distinguishable using the mitochondrial cytochrome b (cytb) gene (see Supplementary File 2 for our analyses on cytb).
As for Botryllus sp. SHJ 5.1 (from Shikine Island, Izu Islands, Japan; in Clade “p” of Figure 3), a formalin-preserved sample was available but the lack of clear diagnostic characters and of the mtCOI sequence necessary to carry out species delimitation analyses led to the assignment of this sample only at genus level (see details in Supplementary File 2).
Updated plates (Supplementary File 3) and morphological descriptions (Supplementary File 4) for four other taxa, originally reported in Nydam et al. (2021) as Botrylloides sp. and Botryllus sp., allowed their description as new species, named here as:
● Botrylloides nigerflavus n. sp. from Panama,
● Botrylloides frankovichi n. sp. from Florida and the Bahamas,
● Botrylloides marikabani n. sp. from Marikaban Island in the Philippines, and
● Botryllus bayanani n. sp. from Catalagan, Marikaban Island, and Puerto Galera in the Philippines.
Species delimitation analyses also support these morphological results (Supplementary Table 2 and blue circle in Figure 3), with the single exception of Botrylloides marikabani n. sp., for which the availability of only two mtCOI sequences (Supplementary Tables 1 and 2) could have hindered its identification as a single new species. Indeed, two distinct OTUs were identified in Botrylloides marikabani by the mtCOI species delimitation analyses (see also the presence of a blue rectangle in Figure 3).
Botrylloides jacksonianum and Botrylloides leptum (inside Clade “o” and “l” of Figure 3, respectively), were already identified as distinct species in the mtCOI botryllid phylogeny of Salonna et al. (2021), but are currently unaccepted as valid species according to the Ascidiacea World Database (Shenkar et al., 2022). Indeed, they were both originally described by Herdman (as Sarcobotrylloides jacksonianum and Botrylloides leptum) (Herdman, 1891; Herdman, 1899), but were then synonymized with Botrylloides leachii (Kott, 1985; Kott, 2005; Shenkar et al., 2022). The mtCOI species delimitation analyses and the phylogenomic trees presented here (Figure 3 and Supplementary File 5) provide further evidence that Botrylloides jacksonianum and Botrylloides leptum are distinct species, both different from B. leachii, and that these Herdman’s species should be resurrected, as previously proposed by Salonna et al. (2021).
The Botrylloides cf. pannosum sample (inside Clade “o”), similar to the original description of Botrylloides pannosum (Herdman, 1899), was also here identified as a distinct species by the species delimitation analyses (Supplementary Table 2 and blue circle in Figure 3) but must be compared to Herdman’s type specimen of Botrylloides pannosum (originally Sarcobotrylloides pannosum) to make an unambiguous taxonomic identification (as proposed in Salonna et al., 2021). If Botrylloides cf. pannosum is indeed Botrylloides pannosum, then Botrylloides pannosum should also be resurrected since, like Botrylloides jacksonianum and Botrylloides leptum, even Botrylloides pannosum was synonymized with Botrylloides leachii (Shenkar et al., 2022). The necessary work to resurrect this additional Herdman’s species is ongoing (M. Ekins, unpublished data).
Overall, the implementation in botryllids of an integrated taxonomy approach, combining morphological as well as nuclear and mitochondrial molecular analyses, allowed us not only to describe six new species but also to identify a number of putative unnamed taxa, for which the examination of additional samples is needed to clarify whether they belong to previously undescribed species. Thus, our results clearly demonstrate the existence of an unexplored hidden diversity within botryllids. This cryptic diversity can be due to factors such as the complexity of morphological analyses, the lack of certain/evident key characters for species discrimination, the paucity of taxonomic experts working in this group, and the historical reliance on morphological characteristics alone for both sample identification and species description.
Phylogenomic tree and the Botryllus-only Clade “a”
Figure 3 shows the phylogenomic tree generated by RAxML from the concatenation of the 177 analyzed nuclear loci, while Supplementary File 5 shows the coalescent summary tree generated by ASTRAL from the 177 single-locus trees. Since the topology of the two trees is almost identical, and in both trees nearly all clades are well supported (bootstrap values = 1 in the ASTRAL tree and 100% in the RAxML tree), we will focus only on the RAxML tree of Figure 3. A Botryllus-only clade (Clade “a”) is the sister clade to the rest of the ingroup, including both Botryllus and Botrylloides species (Figure 3 and Supplementary Figure 5). We were able to examine and describe the morphological characteristics of 4 of the 7 species forming Clade “a”: Botryllus camur (Palomino-Alvarez et al., 2022), Botryllus bonanzus (Palomino-Alvarez et al., 2022), Botryllus primigenus B (Salonna et al., 2021), and Botryllus watanabei n. sp. (Supplementary File 2). All of these species have small zooids with four stigmatal rows, a feature neither found in other botryllids included in this phylogeny nor in the outgroup genus Symplegma, which is characterized by the presence of 8–13 stigmatal rows (Rocha RM, Montesanto F, Nydam M, unpublished data; Mastrototaro et al., 2019). Moreover Botryllus watanabei n. sp. has wide stomach folds, a rectangular stomach shape like Symplegma species (Plate 9g in Supplementary Figure 1), and zooids that exhibit a merging of the 1st and 2nd longitudinal vessels on the left side (Plate 9e in Supplementary Figure 1), which is a Symplegma character. B. primigenus’s species name means “first, primordial, ancestral” in Latin, because it occupies an intermediate position between Botrylloides/Botryllus and other colonial Styelidae with regard to the structure of its systems (Oka, 1928). Notably, our Clade “a” is in agreement with and retains many characteristics considered ancestral in Saito’s botryllid phylogeny (Saito et al., 2001), a phylogeny based on the combination of 18S rDNA sequences and several other features such as life history, reproductive characters, morphology and allorejection mechanisms. In particular, in Saito’s phylogeny (Saito et al., 2001), B. primigenus and all other botryllids with 4 stigmatal rows cluster together and are described as unique in their morphology and life history. So, Saito’s hypothesis is that these species diverged early in the botryllid evolution, although immediately after a group consisting of Botryllus scalaris, Botryllus horridus and Botryllus puniceus. Even in our phylogenomic tree (Figure 3), Botryllus scalaris and Botryllus horridus group together (see Clade “d” in Figure 3), however they do not form the earliest diverging botryllid clade. In general, our tree is in good agreement with Saito’s phylogeny except for the monophyly of the genus Botrylloides, that, remarkably, in our tree become polyphyletic (see red names in Figure 3 and next section). It should be noted that the Botrylloides polyphyly here observed could also be due to our taxon sampling, which was much larger than that of Saito, and in particular to the phylogenetic position of species absent in Saito’s phylogeny since they were described or genetically identified only after 2001, i.e., after the publication date of Saito et al. (2001) (see for example, Clade “p” and the clade Botrylloides conchyliatus/Botrylloides sp. Aust10 in Figure 3 were identified after 2001).
Phylogenomic tree and the status of Botryllus and Botrylloides genera
In Figure 3, both Botrylloides and Botryllus species are found together within the clade “e”, emerging just next to the aforementioned clade “a”, but these two genera do not form separate clusters (see the distribution of the red names in Figure 3, labeling original and reassigned Botrylloides species). Remarkably, the large Clade “j” includes all the analyzed Botrylloides species, except for three species: Botrylloides sp. Aust 10, Botrylloides conchyliatus and Botrylloides crystallinus. The last species forms Clade “i” together with Botryllus sexiens (Plate 5 of Supplementary Files 1 and 2). Based on the original morphological description of Botryllus sexiens (Saito et al., 1981a), this species has zooids arranged in ladder systems with several common cloacal apertures. Therefore, according to the definition of the Botryllus and Botrylloides genera reported in Brunetti (2009), it should be moved to the Botrylloides genus, and renamed to Botrylloides sexiens (hereafter we will refer to this taxon as “Botryllus/Botrylloides” sexiens). This will also keep Clade “i” as Botrylloides only. Given the overall topology of Figure 3 and Supplementary File 5, neither of the two genera is monophyletic, and Botryllus is paraphyletic with respect to Botrylloides. Remarkably, this result was obtained in spite of the fact that the assignment of five specimens was based on the implicit assumption that species of the same genus should cluster together and then that each genus was monophyletic (see previous section “Sample identification reveals a hidden botryllid diversity”: this holds for four Botrylloides and one Botryllus species that were tentatively identified at genus level based just on their positions in the phylogenomic trees, since no formalin-preserved fraction and no suitable mtCOI sequences were available). Overall, the lack of monophyly of both genera aligns with the topologies in previous botryllid phylogenies (Cohen et al., 1998; Nydam et al., 2021).
Assigning a botryllid species to the genus Botryllus or Botrylloides is not easy, also because the definition of these genera has changed over time and some authors have even synonymized them (Monniot, 1988). In general, only few characters have been used to distinguish these genera (Saito et al., 2001 and references therein; Brunetti, 2009 and references therein): the genus Botrylloides has been defined by the presence of zooids arranged in ladder systems, ovary posterior to testis, and embryos developing in a sac-like incubatory pouch forming an outgrowth of the body wall. On the contrary, the genus Botryllus has been defined by the presence of zooids arranged in oval or star-shaped systems, ovary anterior or dorsal to testis, and embryos developing in the peribranchial cavity without a brooding organ. However, Saito already observed inconsistencies in these classification criteria since some botryllid species show features not coinciding with neither the definition of Botryllus nor that of Botrylloides, but have a mixture of the above-mentioned characters (Saito et al., 2001; Okuyama and Saito, 2002). So, he proposed that the definition of the two genera should be amended (Saito et al., 2001). According to Brunetti (2009), who followed the original definition of Milne-Edwards (1841), the two genera differ and should be distinguished by the presence/absence of an atrial siphon and by the consequent structure of the cloaca, which limits the arrangement of the zooids in the systems. Indeed, in Botryllus zooids, the atrial opening is located at the distal end of a conical siphon and its anterior rim extends as a small dorsal tongue, thus allowing only the formation of oval or star-shaped systems. On the contrary, in Botrylloides zooids, there are no atrial siphons and the atrial opening is wide, usually with a broad dorsal lip extending from its anterior rim, thus producing ladder systems. On this basis, it is not surprising that neither Botryllus nor Botrylloides appear monophyletic in our phylogenomic reconstruction (Figure 3 and Supplementary File 5): this lack of monophyly could be biologically significant, just related to the different criteria used for the genus assignment of the new species described over time, or a mixture of both factors. For example, according to the genus definition of Brunetti (2009), both Botryllus sexiens and Botryllus scalaris, described by Saito in 1981, should be moved to the genus Botrylloides: their zooids are indeed arranged in ladder systems with several common cloacal apertures. Therefore, both these species await reassignment to the genus Botrylloides and we labeled them with an asterisk in our phylogenomic tree (Figure 3). The same fate could probably affect even other botryllids, if a detailed re-analysis of the already described species is carried out.
As for the overall topology of the main Botrylloides Clade “j”, in the RAxML tree (Figure 3) Botrylloides saitoi n. sp. is sister to the remaining taxa, however in the ASTRAL coalescent summary tree (Supplementary File 5), the corresponding branch consists of a subclade of three species: Botrylloides saitoi n. sp., Botrylloides jacksonianum, and Botrylloides cf. pannosum (in the RaxML tree, the last two species are joined in Clade “o” that emerges just after the Botrylloides saitoi n. sp. branch). This is the main difference between the RAxML tree from concatenated loci (Figure 3), and the ASTRAL coalescent summary tree from single-locus trees (Supplementary File 5). Other minor differences are reported in Supplementary File 6. The morphological characteristics of Botrylloides saitoi n. sp. place it firmly in the Botrylloides genus (see species description in Plate 6 of Supplementary Files 1, and 2), but its phylogenetic position predicts that it could represent a morphological transition between Botryllus and Botrylloides, alone or together with Botrylloides jacksonianum and Botrylloides cf. pannosum, depending on the considered phylogenomic tree.
Relationships in the clade of the former Botryllus schlosseri complex
In our phylogenetic reconstructions (Figure 3 and Supplementary File 5), Clade “f” encompasses the cryptic species of the former Botryllus schlosseri species complex, i.e.:
● The putative Botryllus renierii, as proposed in Salonna et al. (2021), corresponding to Botryllus schlosseri Clade D sensu Bock et al. (2012);
● The recently described Botryllus gaiae (Brunetti et al., 2020), corresponding to Botryllus schlosseri Clade E sensu Bock et al., 2012; and
● Botryllus schlosseri sensu Brunetti et al. (2017), corresponding to Botryllus schlosseri Clade A sensu Bock et al. (2012). The samples of the latter species are reported in Figure 3 also indicating the subclade to which they belong, according to the phylogeny based on an “Elongated-856nt” mtCOI alignment of Salonna et al. (2021) (see “Subcl A1 to A3” in Figure 3).
The putative Botryllus renierii forms a monophyletic clade that is a sister group to Clade “f” and is well separated from all other samples of the B. schlosseri complex. Noteworthy, Botryllus gaiae is nested in a clade containing all Botryllus schlosseri sensu Brunetti et al. (2017) samples, moreover samples of subclade A2 do not cluster together (Figure 3). On the contrary, in the ASTRAL coalescent summary tree (Supplementary File 5), most of the corresponding nodes (i.e., those in the cluster including Botryllus schlosseri sensu Brunetti et al. (2017) and Botryllus gaiae) have unreliable support values < 0.5. Therefore, based on the RAxML phylogenomic tree neither Botryllus gaiae nor Botryllus schlosseri sensu Brunetti et al. (2017) are monophyletic (Clade “f” in Figure 3), but based on the ASTRAL tree (Supplementary File 5) these relationships are unresolved. Finally, the RAxML result is in disagreement with our mtCOI species delimitation analyses that identify all Botryllus schlosseri sensu Brunetti et al. (2017) as belonging to the same OTU, distinct from the OTU of Botryllus gaiae (Supplementary Table 2; see also blue circles in Figure 3 for Botryllus gaiae and Botryllus schlosseri). A clear distinction between Botryllus gaiae and Botryllus schlosseri sensu Brunetti et al. (2017) was also obtained in all previous mtCOI analyses (Lopez-Legentil et al., 2006; Bock et al., 2012; Yund et al., 2015; Nydam et al., 2021) based on even longer mtCOI alignments (Salonna et al., 2021), as well as by the analysis of the entire mitochondrial genome in several samples (Brunetti et al., 2020).
Overall, these results highlight a possible inconsistency between nuclear and mitochondrial data in the clade of the former Botryllus schlosseri species complex, which appear when only the RAxML result is taken into account. There could be several explanations for this inconsistency:
a) The anchored hybrid enrichment here used is designed for capturing orthologous loci characterized by conserved anchor regions plus neighboring variable flanking regions, so in principle it is able to resolve both shallow and deep taxonomic relationships (Hamilton et al., 2016). However, it is still possible that our dataset does not include enough variable informative loci to accurately resolve the short branches inside the former Botryllus schlosseri species complex. This explanation is supported by the inconsistency of the results of the RAxML compared to the ASTRAL analyses and by the finding that the nuclear locus 18S rDNA recovered distinct well-supported cryptic species in the Botryllus schlosseri species complex (Bock et al., 2012), and between Botryllus schlosseri and Botryllus gaiae (Nydam et al., 2017b).
b) We cannot exclude the possibility that the inconsistency between the nuclear and the mitochondrial phylogenies (with lack of monophyly of the former cryptic species in our nuclear RAxML tree) could be due to introgression and hybridization between the original cryptic species of the species complex. However, no evidence of gene flow was found between B. schlosseri and B. gaiae at seven microsatellite loci (Bock et al., 2012), and one of the two B. gaiae samples in our nuclear tree came from a population (Falmouth, UK) from which the microsatellite data were obtained.
In conclusion, the current data make it impossible to determine whether the nuclear or mitochondrial phylogeny represents the true evolutionary history of the former Botryllus schlosseri species complex. This issue could be solved only by the parallel analyses of a larger dataset of complete mitochondrial genomes including representative of all cryptic species of this taxon, and of a larger dataset of nuclear loci, ad hoc designed for the evolutionary distances present within the former Botryllus schlosseri species complex.
Mapping mtCOI species delimitation on the phylogenomic tree
The phylogenomic tree of Figure 3 was assembled to cover a broad range of the genetic diversity within the Botrylloides/Botryllus tree, so it does not allow the species delimitation analyses because it contains insufficient information on the intra-species variability. Therefore, our species delimitation results based on mtCOI sequences (Supplementary Table 2) were mapped on this nuclear phylogenomic tree in order to identify species-level clades. The cross-referencing of these results is particularly important for closely related taxa, i.e., for the sister taxa/clades showing very short internal branch lengths in Figure 3.
In particular, the results of the mtCOI species delimitation analyses (Supplementary Table 2) were summarized in Figure 3 using a blue circle to indicate species consisting of a single OTU and a blue rectangle to indicate species consisting of multiple OTUs. In most cases, the nuclear and the mtCOI data used for the phylogenomic and the species delimitation analyses, respectively, gave congruent results. For example, the two Botryllus primigenus samples 1S and 2S, already identified as distinct OTUs in Salonna et al. (2021), were not found to be sister taxa but were placed in separate clades by the nuclear data. In agreement with this result, they were identified as two distinct OTUs by the mtCOI species delimitation analyses (Supplementary Table 2). Therefore, these two samples should be considered as belonging to two different species, named in Figure 3 as “Botryllus primigenus A” for sample 1S and “Botryllus primigenus B” for sample 2S, although they were morphologically identified and provided by Saito Y. as belonging to the same Botryllus primigenus species (personal communication). A partial agreement between nuclear and mtCOI data can be seen in Clade “f”, which includes the former Botryllus schlosseri species complex (see above section).
A total of eight species are each identified as consisting of multiple OTUs by the mtCOI species delimitation analyses (see species with a blue rectangle in Figure 3), suggesting that additional molecular samples/data are needed to better clarify the taxonomic status of these species or indicating inconsistencies between molecular and morphological data. Some of these cases were already discussed in Nydam et al. (2021) (for Botrylloides marikabani n. sp.) and in Palomino-Alvarez et al. (2022) (for Botrylloides unamensis and Botrylloides catalitinae), while for Botrylloides violaceus the lack of morphological information on the samples present in the tree prevents us from formulating hypotheses explaining this result.
Within clade “n”, all our Botrylloides diegensis samples forms a single OTU together with the Botrylloides leachii samples published in Salonna et al. (2021) (Supplementary Table 2): this is in agreement with Viard’s proposal that all published mtCOI sequences of B. leachii should be re-assigned to Botrylloides diegensis (Viard et al., 2019). However, the samples assigned to Botrylloides diegensis in Viard et al. (2019) were described only for the colony color pattern, without details on the anatomical traits discriminating Botrylloides leachii from Botrylloides diegensis (i.e., the number of rows of stigmata, the presence/absence of muscle in the branchial sac, and the number of stomach folds (Brunetti and Mastrototaro, 2017)). Since the absence of these morphological data prevent the evaluation of a possible case of species synonymy, in Figure 3 we have labeled this clade/OTU as “Botrylloides diegensis/leachii”. Combined morphological and molecular analyses, performed on exactly the same samples, will be necessary to resolve this question.
Additional comments on the mapping of mtCOI species delimitation results on the phylogenomic tree are reported in Supplementary File 6.
Phylogenomic implications for the evolution of allorecognition
Our goal in building the botryllid phylogenomic tree was to document the evolution of allorecognition in the Botrylloides and Botryllus genera. Allorecognition R-types were therefore mapped on Figure 3 and Supplementary File 5, which was rooted with Symplegma, i.e., the sister genus to the Botrylloides/Botryllus group. Symplegma also has zooids connected via a vascular system with ampullae (Shirae et al., 1999) and can undergo allorecognition. The rejection reaction has so far been studied in a single Symplegma species, S. reptans, where it has been found that some colonies exhibit only R-Type B and other colonies only R-Type E rejection reactions (Shirae et al., 1999). However, the R-Type E of S. reptans differs from R-Type E of B. scalaris, since the rejection process is mainly mediated by the activation of morula cells in S. reptans and by phagocytes in B. scalaris (Shirae et al., 1999). Thus, in these two species, the R-Type E is morphologically similar, since it involves the same allorecognition site, but is characterized by differences in the hemocyte behavior and thus probably in the underlying molecular processes. Remarkably, morula cells, not phagocytes, have been found to be principal “effector cells” in the rejection reaction in the few other botryllids (all R-Type A) in which the hemocyte histochemistry of this process has been investigated (Shirae et al., 1999). With the caveat that this inference is based on a single outgroup species, we can infer that at least R-Type B and an R-Type “E-like” process are ancestral to the entire botryllid group.
Cohen et al. (1998) showed that the allorecognition response involving the most internal tissues (R-Type E) is the ancestral allorecognition condition, thus providing evidence for an evolutionary progression of the allorecognition system from R-Type E to R-Type D and then to R-Type A (Cohen et al., 1998 and Figure 2A). Our phylogenomic trees do not support this finding (Figure 3 and Supplementary File 5) and the application of an ancestral character state reconstruction program does not provide conclusive results (Supplementary Files 7–9 and their detailed description in Supplementary File 6). Indeed, in the ancestral character state reconstructions, several clades show more than one equiparsimonious state, and the results vary considerably depending on the R-Type assigned to the outgroup species, which is not definitely known (indeed Symplegma shows at least two different R-Types). Our reconstructions should also be viewed with several caveats since: 1) parsimony is not the only framework in which to conduct these analyses; 2) 73–75% of our specimens have an unknown R-Type, which decreases the robustness of the overall analysis; and 3) the nested position of B. sexiens in our phylogeny, inside Clade “h”, strongly affects the overall ancestral state reconstructions since its R-type is only putative (it should be R-Type E) and it is the only “not R-Type A” included in the large Clade “h”. In our opinion, the preponderance of unknown R-Types and the uncertainty of B. sexiens’ R-Type are the main reasons leading to view our ancestral reconstructions as provisional.
Our analyses differ from those of Cohen in several aspects, including the usage of an ancestral character state reconstruction software strictly following the parsimony criterion and the outgroup choice. In particular, this last factor can strongly affect the tree topology. Compared to Cohen et al. (1998), our outgroup was more accurately selected since it is the genus phylogenetically nearest to botryllids, and also shows allorejection reactions similar to botryllids. Our overall picture of the evolution of the rejection reaction in botryllids is much more complex but still not definitive. First of all, we have insufficient data to define the ancestral R-Type in the Symplegma outgroup, since the rejection reaction was studied only in one species (Symplegma reptans) where both R-Type B and an R-Type “E-like” were observed (Shirae et al., 1999). So, probably many other R-Types are present in the Symplegma genus outgroup. Thus, without clear and complete data on the R-Type of the outgroups, several alternative scenarios can be hypothesized for the botryllid allorecognition evolution. For example, R-Type D or E can be considered as the ancestral status of all botryllids if the Symplegma R-Types data are not taken into account (Supplementary File 7), while R-Type B, D or E can be the ancestral status of all botryllids if one of the known Symplegma R-Types is accepted as ancestral (Supplementary Files 8 and 9). Although the R-Type is missing for many species of our phylogeny, the R-Type A species are all clustered together and certainly evolved later, in Clade “n” at the latest (Supplementary Files 7–9). Finally, to understand the evolution of allorejection in botryllids, this process needs to be investigated also at the level of hemocyte behavior (i.e., cells that recognize self–non-self and effector cells), not only of allorejection sites (Shirae et al., 1999), in order to better classify the different R-Types.
Although we do not obtain conclusive results, our data provide interesting observations. Clade “d” includes all available samples of the former Botryllus schlosseri species complex and it displays either R-Type B or R-Type C phenotypes, with the R-Type B observed in populations from California (Pacific Ocean) and from Venice (Mediterranean Sea), and the R-Type C observed in populations from Massachusetts (Atlantic Ocean) (Table 1). At the time the Botryllus schlosseri rejection experiments were carried out (Boyd et al., 1990), its status as species complex was unknown, so it is now impossible to unambiguously associate a specific R-Type to each cryptic species without repeating the allorecognition experiments on genetically characterized colonies. However, based on sample geographic origin, we have mapped R-Type B at the level of our sample coming from California (i.e., “Botryllus schlosseri, unk. clade” in Figure 3) and from Venice (i.e., “Botryllus schlosseri, NeoA and NeoD in Figure 3), while we have not associated R-Type C to any specific Botryllus schlosseri sample, since in our tree there are no Botryllus schlosseri samples coming from Massachusetts. We have also tentatively mapped R-Type C at the level of the basal node including of the Botryllus schlosseri species complex, since California and Massachusetts samples analyzed by Boyd et al. (1990) all certainly belong to the Botryllus schlosseri species complex. Moreover, we hypothesize that both R-Type B and R-Type C are present in Botryllus schlosseri sensu Brunetti et al. (2017), i.e., in clade A sensu Bock et al. (2012), since current cryptic species distribution data shows that Botryllus schlosseri sensu Brunetti et al. (2017) is the only species widespread worldwide, while Botryllus gaiae (i.e., the former cryptic clade E) is present only in the European waters (including the Mediterranean Sea), and the other cryptic species are geographically restricted to a few localities (see Brunetti et al., 2017; Nydam et al., 2017a; Brunetti et al., 2020 and references therein). Botryllus scalaris (which, as previously discussed, should be moved to the genus Botrylloides) is the only one of three known R-Type E species (Table 1) present in our tree, and is found in Clade “d”, so close to the Botryllus schlosseri species complex.
Clade “h” includes all the currently known R-Type A species (Table 1), however we do not know if this clade includes even other R-Types, as many of its species have not been analyzed for R-Types. In support of the presence of species other than R-Type A in Clade “h”, Botryllus sexiens (that we proposed to move into the Botrylloides genus) is thought to be R-Types E (Cohen et al., 1998). It would be interesting to study the R-Types in Clade “p”, which is a small Botryllus clade in the large Clade “h” consisting mostly of Botrylloides.
Botryllus R-Types so far known are B, C, D, and E, while Botrylloides are only R-Type A, but they will also include R-Type E if our proposal to reassign both Botryllus sexiens and Botryllus scalaris to the genus Botrylloides is considered valid.
There are six genes present in the Botrylloides and Botryllus genera that have characteristics consistent with a role in allorecognition: two highly polymorphic genes (fuhcsec and fuhctm) (Nydam et al., 2013a), two genes encoding for putative receptors (fester and uncle fester) (Nyholm et al., 2006; McKitrick et al., 2011), and two genes which may be involved in generating specificity (Hsp40-L and BHF) (Nydam et al., 2013b; Voskoboynik et al., 2013). Since uncle fester is responsible for the rejection response (McKitrick et al., 2011), we predict that a different expression pattern of this gene could be responsible for the different R-Type reactions. In particular, the presence of this protein only on circulatory cells in R-Type E species would explain the complete membrane fusion of colony circulatory systems prior to rejection. On the contrary, R-Type A–D reactions are likely the result of expressing uncle fester on more external tissues (e.g., on the surface of the tunic cuticle, in the subcuticular region, within the tunic matrix, or at the surface of the ampullar epithelium). This hypothesis needs to be tested by tissue-specific transcriptome analyses.
The results of xenorecognition experiments, wherein colonies from different species interact with each other (Saito, 2003), can also be mapped onto our phylogenomic tree. Unlike the allorejection reaction, in xenorecognition the rejection is the default, and the fusion response is an interruption of that process. There are three outcomes from xenorecognition experiments, which can also vary depending on the conditions (contact at growing edges, at cut surfaces, or blood plasma injection): 1) both species initiated a rejection response, 2) neither species initiated a rejection response, 3) one species initiated a rejection response and the other did not (Saito, 2003). If the loci responsible for initiating the rejection response are species-specific, then the more closely related species pairs would be predicted to initiate rejection responses, while the more distantly related species pairs would not. The three participant species in the Saito’s xenorecognition experiments were Botrylloides simodensis (R-Type A), Botryllus primigenus (R-Type D), and Botryllus scalaris (R-Type E) (Saito, 2003). Taking into account the results of both growing-edge and cut surface contacts, B. primigenus (R-Type D) initiated a rejection reaction with both B. simodensis (R-Type A) and B. scalaris (R-Type E) (Saito, 2003). B. simodensis (R-Type A) initiated a rejection reaction with both B. primigenus (R-Type D) and B. scalaris (R-Type E) (Saito, 2003). In contrast, B. scalaris (R-Type E) did not react with either the R-Type A or the R-Type D species in both conditions (Saito, 2003). In the Cohen et al., 1998 phylogeny (Figure 2A), the R-Type E species is sister to the rest of the species and is referred in that paper as emerging early in the tree, so the earlier evolving species could not initiate a rejection response with later evolving species, but the later evolving R-Type A and the R-Type D species had the capacity to initiate a rejection response with the earlier evolving R-Type E species. In the current phylogeny (Figure 3 and Supplementary File 5), the R-Type E species B. scalaris is sister group to the former B. schlosseri complex, including R-Type B and putative R-Type C species (see Figure 3B; Supplementary File 5B and Clade “d” in Figure 3A and Supplementary File 5A) but, to our knowledge, rejection reactions between R-Type B/C and R-Type E have not been tested. However, it seems that the loci responsible for xenorejection initiation in R-Type E may have evolved in such a way as to be incompatible with R-Type A and R-Type D. This is also in accordance with a greater separation between R-Type E and all other R-Types, due for example to the major involvement of phagocytes instead of morula cells in the R-Type E allorejection reaction (Shirae et al., 1999).
Xenorecognition experiments within R-Type A species (B. fuscus, B. lentus, B. simodensis, and B. violaceus) have also been conducted (Hirose et al., 2002). The results of a contact at colony growing edges do not correlate with phylogenetic position. Indeed, B. violaceus reacts with B. fuscus with an intense hemolytic reaction but not with B. lentus or B. simodensis (Hirose et al., 2002), although B. violaceus is sister to the clade that includes the other three species (Figure 3 and Supplementary File 5). B. fuscus and B. lentus are sister species in our phylogeny, but they do not react the same way when the growing edges of their colonies contact either B. simodensis or B. violaceus (Hirose et al., 2002).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical review and approval was not required for the study on animals in accordance with the local legislation and institutional requirements.
Author contributions
MN: Project management, sample contribution, DNA barcoding, species delimitation analyses, morphological analyses, manuscript writing. AL: Project management on the genomic work. EL: Project management on the genomic work. KZ: Alignment and assembly of genomic reads, tree creation. CSC: Sample contribution. LP-A: Sample contribution. CG: Sample contribution, manuscript writing. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. MN was funded by a Luce Initiative for Asian Studies and the Environment Grant awarded to Centre College, and by Soka University of America. CG acknowledges the Molecular Biodiversity Laboratory of the Italian node of Lifewatch (CNR, Consiglio Nazionale delle Ricerche, Italy), Elixir-IT (the European Research Infrastructure for Life Science), and CNRBiOmics (Infrastructural project PIR01_00017 of MUR – Ministero dell’Università e della Ricerca, Italy). Posgrado en Ciencias del Mar y Limnología, UNAM, and Consejo Nacional de Ciencia y Tecnología (2018-000012-01NACF-08376, CVU 447073) supported LP-A. Grants from the Harte Charitable Foundation and CONABIO-NE018 to Nuno Simoes supported the samples obtained from the Gulf of Mexico and the Caribbean by LP-A.
Acknowledgments
We would like to thank the following people for assistance with sample collection: Sylvain Agostini, Erin Grey-Avis, Tony De Tomaso, Ben Harvey, Morgan Hawkins, Kazuo Inaba, Koetsu Kon, Yukinobu Isowa, Toru Miura, Hitoshi Sawada, Maki Shirae-Kurabayashi, Lauren Stefaniak. We thank Loriano Ballarin and Euichi Hirose for helpful discussions on the details of the rejection reaction in botryllids. We thank Alberto Guerra for drawing Figure 1. The insightful comments of three reviewers improved the manuscript. We also thank the Italian MEDITS survey program, the Steinhardt Museum of Natural History (National Center for Biodiversity Studies, Israel), the Queensland Museum (Australia) and the lamented Yasunori Saito for kindly providing the ascidian samples previously analyzed in Salonna et al. (2021). We acknowledge The Philippines’ Bureau of Fisheries and Aquatic Resources and the National Fisheries Research and Development Institute for their role in the 2014–2016 Verde Island Passage (VIP) Expedition. All B. marikabani specimens were collected during the NSF Biotic Survey VIP Expedition under Gratuitous Permit # GP-0077-14 from the municipalities of Puerto Galera and Batangas.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1214191/full#supplementary-material
References
Atsumi M. O., Saito Y. (2011). Studies on Japanese botryllid ascidians. V. A new species of the genus Botrylloides very similar to Botrylloides simodensis in morphology. Zoological Sci. 28, 532–542. doi: 10.2108/zsj.28.532
Ballarin L., Karahan A., Salvetti A., Rossi L., Manni L., Rinkevich B., et al. (2021). Stem cells and innate immunity in aquatic invertebrates: bridging two seemingly disparate disciplines for new discoveries in biology. Front. Immunol. 12. doi: 10.3389/fimmu.2021.688106
Ben-Shlomo R., Reem E., Douek J., Rinkevich B. (2010). Population genetics of the invasive ascidian Botryllus schlosseri from South American coasts. Mar. Ecol. Prog. Ser. 412, 85–92. doi: 10.3354/meps08688
Berrill N. J. (1950). The Tunicata with an account of the British species. Ray Society. (London: Ray Society, Johnson Reprints).
Blanchoud S., Rinkevich B., Wilson M. J. (2018). Whole-body regeneration in the colonial tunicate Botrylloides leachii. Mar. Organisms as Model. Syst. Biol. Med. 65, 337–355. doi: 10.1007/978-3-319-92486-1_16
Bock D. G., MacIsaac H. J., Cristescu M. E. (2012). Multilocus genetic analyses differentiate between widespread and spatially restricted cryptic species in a model ascidian. Proc. R. Soc. B: Biol. Sci. 279, 2377–2385. doi: 10.1098/rspb.2011.2610
Boyd H. C., Weissman I. L., Saito Y. (1990). Morphologic and genetic verification that Monterey Botryllus and Woods Hole Botryllus are the same species. Biol. Bull. 178, 239–250. doi: 10.2307/1541825
Brunetti R. (2009). Botryllid species (Tunicata, Ascidiacea) from the Mediterranean coast of Israel, with some considerations on the systematics of Botryllinae. Zootaxa 2289, 18–32. doi: 10.11646/zootaxa.2289.1.2
Brunetti R., Griggio F., Mastrototaro F., Gasparini F., Gissi C. (2020). Toward a resolution of the cosmopolitan Botryllus schlosseri species complex (Ascidiacea, Styelidae): mitogenomics and morphology of clade E (Botryllus gaiae). Zoological J. Linn. Soc. 190, 1175–1192. doi: 10.1093/zoolinnean/zlaa023
Brunetti R., Mastrototaro F. (2017). Fauna d’Italia. Ediz. Vol. 2: Ascidiacea of the European Waters. (Milan, Italy: Edagricole-New Business Media II). Vol. 51.
Brunetti R., Mastrototaro F. (2017). “Ascidiacea of the European waters,” in Edagricole - New Business Media II.
Buss L. (1982). Somatic cell parasitism and the evolution of somatic tissue compatibility. Proc. Natl. Acad. Sci. 79, 5337–5341. doi: 10.1073/pnas.79.17.5337
Cohen C. S., Saito Y., Weissman I. L. (1998). Evolution of allorecognition in botryllid ascidians inferred from a molecular phylogeny. Evolution 52, 746–756. doi: 10.1111/j.1558-5646.1998.tb03699.x
De Tomaso A., Nyholm S. V., Palmeri K. J., Ishizuka K. J., Ludington W. B., Mitchel K., et al. (2005). Isolation and characterization of a protochordate histocompatibility locus. Nature 438, 454–459. doi: 10.1038/nature04150
Ducasse J., Ung V., Lecointre G., Miralles A. (2020). LIMES: A tool for comparing species partitions. Bioinformatics 36, 2282–2283. doi: 10.1093/bioinformatics/btz911
Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299.
Hamilton C. A., Lemmon A. R., Lemmon E. M., Bond J. E. (2016). Expanding anchored hybrid enrichment to resolve both deep and shallow relationships within the spider tree of life. BMC Evolutionary Biol. 16, 212. doi: 10.1186/s12862-016-0769-y
Herdman W. A. (1891). A revised classification of the Tunicata, with definitions of the orders, suborders, families, subfamilies, and genera, and analytical keys to the species. J. Linn. Soc. London 23, 558–652.
Herdman W. A. (1899). Descriptive catalogue of the Tunicata in the Australian Museum (Sydney N.S.W. T: Dobb and Company).
Hirose E., Saito Y., Watanabe H. (1988). A new type of manifestation of colony specificity in the compound ascidian, Botrylloides violaceus Oka. Biol. Bull. 175, 240–245. doi: 10.2307/1541564
Hirose E., Saito Y., Watanabe H. (1990). Allogeneic rejection induced by cut surface contact in the compound ascidian, Botrylloides simodensis. Invertebrate Reprod. Dev. 17, 159–164. doi: 10.1080/07924259.1990.9672105
Hirose E., Shirae M., Saito Y. (2002). Colony specificity in the xenogeneic combinations among four Botrylloides species (Urochordata, Ascidiacea). Zoological Sci. 19, 747–753. doi: 10.2108/zsj.19.747
Kott P. (1985). The Australian ascidiacea. Part I, phlebobranchia and Stolidobranchia. Memoirs Queensland Museum 23, 1–440.
Kott P. (2005). Catalogue of Tunicata in Australian waters. Australian Biological Resources Study. (Queensland Museum, Brisbane: Australia Department of the Environment and Heritage).
Lauzon R. J., Brown C., Kerr L., Tiozzo S. (2013). Phagocyte dynamics in a highly regenerative urochordate: insights into development and host defense. Dev. Biol. 374, 357–373. doi: 10.1016/j.ydbio.2012.11.006
Lopez-Legentil S., Turon X., Planes S. (2006). Genetic structure of the star sea squirt, Botryllus schlosseri, introduced in southern European harbours. Mol. Ecol. 15, 3957–3967. doi: 10.1111/j.1365-294x.2006.03087.x
Maddison W. P., Maddison D. R. (2023) Mesquite: a modular system for evolutionary analysis. Version 3.81. Available at: http://www.mesquiteproject.org.
Mastrototaro F., Montesanto F., Salonna M., Grieco F., Trainito E., Chimienti G., et al. (2019). Hitch-hikers of the sea: concurrent morphological and molecular identification of Symplegma brakenhielmi (Tunicata: Ascidiacea) in the western Mediterranean Sea. Mediterr. Mar. Sci. 20, 197–207. doi: 10.12681/mms.19390
McKitrick T. R., Muscat C. C., Pierce J. D., Bhattacharya D., De Tomaso A. W. (2011). Allorecognition in a basal chordate consists of independent activating and inhibitory pathways. Immunity 34, 616–626. doi: 10.1016/j.immuni.2011.01.019
Miller M. A., Pfeiffer W., Schwartz T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, 14 Nov. 2010 (New York City). 1–8. doi: 10.1109/gce.2010.5676129
Milne-Edwards H. (1841). Observations sur les ascidies composées des côtes de la Manche. Chez Fortin-Masson Cie (Paris, France).
Monniot C. (1988). Ascidies de nouvelle-caledonie. IV. Styelidae. Bull. Natl. Museum Natural History Paris 10, 163–196. doi: 10.5962/p.292203
Nourizadeh S., Kassmer S., Rodriguez D., Hiebert L. S., De Tomaso A. W. (2021). Whole body regeneration and developmental competition in two botryllid ascidians. Evodevo 12, 15. doi: 10.1186/s13227-021-00185-y
Nydam M. L., Giesbrecht K. B., Stephenson E. E. (2017a). Origin and dispersal history of two colonial ascidian clades in the Botryllus schlosseri species complex. PloS One 12, e0169944. doi: 10.1371/journal.pone.0169944
Nydam M. L., Hoang T. A., Shanley K. M., De Tomaso A. W. (2013b). Molecular evolution of a polymorphic HSP40-like protein encoded in the histocompatibility locus of an invertebrate chordate. Dev. Comp. Immunol. 41, 128–136. doi: 10.1016/j.dci.2013.03.004
Nydam M. L., Lemmon A. R., Cherry J. R., Kortyna M. L., Clancy D. L., Hernandez C., et al. (2021). Phylogenomic and morphological relationships among the botryllid ascidians (Subphylum Tunicata, Class Ascidiacea, Family Styelidae). Sci. Rep. 11, 1–13. doi: 10.1038/s41598-021-87255-2
Nydam M. L., Neutschil N., Sanders E., Langenbacher A., Lewis D. L., Marimuthu A., et al. (2013a). The candidate histocompatibility locus of a basal chordate encodes two highly polymorphic proteins. PLoS One 8, e65980. doi: 10.1371/journal.pone.0065980
Nydam M. L., Stephenson E. E., Waldman C. E., De Tomaso A. W. (2017b). Balancing selection on allorecognition genes in the colonial ascidian Botryllus schlosseri. Dev. Comp. Immunol. 69, 60–74. doi: 10.1016/j.dci.2016.12.006
Nyholm S. V., Passegue E., Ludington W., Voskoboynik A., Mitchel K., Weissman I. L., et al. (2006). fester, a candidate allorecognition receptor from a primitive chordate. Immunity 25, 163–173. doi: 10.1016/j.immuni.2006.04.011
Oka A. (1928). Ueber eine merkwürdige Botryllus-Art, B. primigenus nov. sp. Proc. Imperial Acad. 4, 303–305.
Okuyama M., Saito Y. (2001). Studies on Japanese botryllid ascidians. I. A new species of the genus Botryllus from the Izu Islands. Zoological Sci. 18, 261–267. doi: 10.2108/zsj.18.261
Okuyama M., Saito Y. (2002). Studies on Japanese botryllid ascidians. II. A new species of the genus Botryllus from the vicinity of Shimoda. Zoological Sci. 19, 809–815. doi: 10.2108/zsj.19.809
Okuyama M., Saito Y., Hirose E. (2002). Fusion between incompatible colonies of a viviparous ascidian. Botrylloides lentus. Invertebrate Biol. 121, 163–169. doi: 10.1111/j.1744-7410.2002.tb00057.x
Palomino-Alvarez L. A., Nydam M. L., Rocha R. M., Simoes N. (2022). New Botrylloides, Botryllus and Symplegma (Ascidiacea: Styelidae) in coral reefs of the southern Gulf of Mexico and Mexican Caribbean Sea. MDPI Diversity 14, 977. doi: 10.3390/d14110977
Puillandre N., Brouillet S., Achaz G. (2021). ASAP: assemble species by automatic partitioning. Mol. Ecol. Resour. 21, 609–620. doi: 10.1111/1755-0998.13281
Puillandre N., Lambert A., Brouillet S., Achaz G. (2012). ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 21, 1864–1877. doi: 10.1111/j.1365-294x.2011.05239.x
Sabbadin A., Zaniolo G., Ballarin L. (1992). Genetic and cytological aspects of histocompatibility in ascidians. Bollettino della Società zoologica italiana 59, 167–173. doi: 10.1080/11250009209386665
Saito Y. (2003). Xenogeneic rejection among three Botryllids (compound ascidians). Zoological Sci. 20, 581–589. doi: 10.2108/zsj.20.581
Saito Y., Hirose E., Watanabe H. (1994). Allorecognition in compound ascidians. Int. J. Dev. Biol. 38, 237–247.
Saito Y., Mukai H., Watanabe H. (1981a). Studies of Japanese compound styelid ascidians. I. Two new species of Botryllus from the vicinity of Shimoda. Publications Seto Mar. Biol. Lab. 26, 347–355. doi: 10.5134/176033
Saito Y., Mukai H., Watanabe H. (1981b). Studies of Japanese compound styelid ascidians. II. A new species of the genus Botrylloides and redescription of Botrylloides violaceus. Publications Seto Mar. Biol. Lab. 26, 357–368. doi: 10.5134/176032
Saito Y., Nagasawa N. (2003). Studies on Japanese botryllid ascidians. III. A new species of the genus Botryllus with a vivid colony color from the vicinity of Shimoda. Zoological Sci. 20, 765–771. doi: 10.2108/zsj.20.765
Saito Y., Okuyama M. (2003). Studies on Japanese botryllid ascidians. IV. A new species of the genus Botryllus with a unique colony shape, from the vicinity of Shimoda. Zoological Sci. 20, 1153–1161. doi: 10.2108/zsj.20.1153
Saito Y., Shirae M., Okuyama M., Cohen C. S. (2001). “Phylogeny of botryllid ascidians,” in The Biology of Ascidians. Eds. Sawada H., Yokosawa H., Lambert C. C. (Berlin/Heidelberg: Springer), 315–320. doi: 10.1007/978-4-431-66982-1_50
Saito Y., Watanabe H. (1982). Colony specificity in the compound ascidian, Botryllus scalaris. Proc. Japan Acad. Ser. B, 58, 105–108. doi: 10.2183/pjab.58.105
Saito Y., Watanabe H. (1985). Studies on Japanese compound styelid ascidians-IV. Three new species of the genus Botrylloides from the vicinity of Shimoda. Publications Seto Mar. Biol. Lab. 30, 227–240. doi: 10.5134/176112
Salonna M., Gasparini F., Huchon D., Montesanto F., Haddas-Sasson M., Ekins M., et al. (2021). An elongated COI fragment to discriminate botryllid species and as an improved ascidian DNA barcode. Sci. Rep. 11, 1–19. doi: 10.1101/2021.01.20.427448
Scofield V. L., Nagashima L. S. (1983). Morphology and genetics of rejection reactions between oozoids from the tunicate Botryllus schlosseri. Biol. Bull. 165, 733–744. doi: 10.2307/1541475
Shenkar N., Gittenberger A., Lambert G., Rius M., Rocha R. M., Swalla B. J., et al. (2022) Ascidiacea World Database. Available at: https://www.marinespecies.org/ascidiacea (Accessed 2022-10-24).
Shirae M., Hirose E., Saito Y. (1999). Behavior of hemocytes in the allorecognition reaction in two compound ascidians, Botryllus scalaris and Symplegma reptans. Biol. Bull. 197, 188–197. doi: 10.2307/1542614
Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stefaniak L., Lambert G., Gittenberger A., Zhang H., Lin S., Whitlatch R. B. (2009). Genetic conspecificity of the worldwide populations of Didemnum vexillum Kott 2002. Aquat. Invasions 4, 29–44. doi: 10.3391/ai.2009.4.1.3
Tanaka K., Watanabe H. (1973). Allogeneic inhibition in a compound ascidian, Botryllus primigenus Oka. I. Processes and features of "Nonfusion" reaction. Cell. Immunol. 7, 410–426. doi: 10.1016/0008-8749(73)90206-2
Viard F., Roby C., Turon X., Bouchemousse S., Bishop J. (2019). Cryptic diversity and database errors challenge non-indigenous species surveys: an illustration with Botrylloides spp. in the English Channel and Mediterranean Sea. Front. Mar. Sci. 6, 615. doi: 10.3389/fmars.2019.00615
Voskoboynik A., Newman A. M., Corey D. M., Sahoo D., Pushkarev D., Neff N. F., et al. (2013). Identification of a colonial chordate histocompatibility gene. Science 341, 384–387. doi: 10.1126/science.1238036
Voskoboynik A., Weissman I. L. (2015). Botryllus schlosseri, an emerging model for the study of aging, stem cells, and mechanisms of regeneration. Invertebrate Reprod. Dev. 59, 33–38. doi: 10.1080/07924259.2014.944673
Yund P. O., Collins C., Johnson S. L. (2015). Evidence of a native northwest Atlantic COI haplotype clade in the cryptogenic colonial ascidian Botryllus schlosseri. Biol. Bull. 228, 201–216. doi: 10.1086/bblv228n3p201
Zaniolo G., Ballarin L. (2001). “Colony specificity in botrylloides leachi (Savigny): preliminary reports,” in The Biology of Ascidians. Eds. Sawada H., Yokosawa H., Lambert C. C. (Tokyo: Springer). doi: 10.1007/978-4-431-66982-1_65
Zaniolo G., Manni L., Ballarin L. (2006). Colony specificity in Botrylloides leachi. I. Morphological aspects. ISJ-Invertebrate Survival J. 3, 125–136.
Keywords: Botryllus, Botrylloides, ascidian, allorecognition, histocompatibility, rejection
Citation: Nydam ML, Lemmon AR, Lemmon EM, Ziegler K, Cohen CS, Palomino-Alvarez LA and Gissi C (2023) Phylogenomics and systematics of botryllid ascidians, and implications for the evolution of allorecognition. Front. Ecol. Evol. 11:1214191. doi: 10.3389/fevo.2023.1214191
Received: 11 May 2023; Accepted: 09 November 2023;
Published: 04 December 2023.
Edited by:
Paolo Sordino, Anton Dohrn Zoological Station Naples, ItalyReviewed by:
Tony De Tomaso, University of California, Santa Barbara, United StatesPaul Simion, University of Namur, Belgium
David Osca, University of Las Palmas de Gran Canaria, Spain
Copyright © 2023 Nydam, Lemmon, Lemmon, Ziegler, Cohen, Palomino-Alvarez and Gissi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marie L. Nydam, mnydam@soka.edu
 Marie L. Nydam
Marie L. Nydam Alan R. Lemmon
Alan R. Lemmon Emily M. Lemmon
Emily M. Lemmon Kevin Ziegler
Kevin Ziegler C. Sarah Cohen
C. Sarah Cohen Lilian A. Palomino-Alvarez
Lilian A. Palomino-Alvarez Carmela Gissi
Carmela Gissi