Abstract
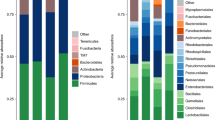
Microbiota perform vital functions for their mammalian hosts, making them potential drivers of host evolution. Understanding effects of environmental factors and host characteristics on the composition and biodiversity of the microbiota may provide novel insights into the origin and maintenance of these symbiotic relationships. Our goals were to (1) characterize biodiversity of oral and rectal microbiota in bats from Puerto Rico; and (2) determine the effects of geographic location and host characteristics on that biodiversity. We collected bats and their microbiota from three sites, and used four metrics (species richness, Shannon diversity, Camargo evenness, Berger-Parker dominance) to characterize biodiversity. We quantified the relative importance of site, host sex, host species-identity, and host foraging-guild on biodiversity of the microbiota. Microbe biodiversity was highly variable among conspecifics. Geographical location exhibited consistent effects, whereas host sex did not. Within each host guild, host species exhibited consistent differences in biodiversity of oral microbiota and of rectal microbiota. Oral microbe biodiversity was indistinguishable between guilds, whereas rectal microbe biodiversity was significantly greater in carnivores than in herbivores. The high intraspecific and spatial variation in microbe biodiversity necessitate a relatively large number of samples to statistically isolate the effects of environmental or host characteristics on the microbiota. Species-specific biodiversity of oral microbiota suggests these communities are structured by direct interactions with the host immune system via epithelial receptors. In contrast, the number of microbial taxa that a host gut supports may be driven by host diet-diversity or composition.


Similar content being viewed by others
Data Availability
Sequencing data of V4 region of the 16S rRNA gene has been deposited in the NCBI Short Read Archive database under BioProject PRJNA602518 and accession numbers SRX7587313-7587772.
Code Availability
Not applicable.
References
Groussin M, Maze F, Sanders JG, Smillie CS, Lavergne S, Thuiller W, Alm EJ (2017) Unraveling the processes shaping mammalian gut microbiomes over evolutionary time. Nat Commun 8:14319
Ingala MR, Simmons NB, Perkins SL (2018) Bats are an untapped system for understanding microbiome evolution in mammals. mSphere 3:e00397-18–8
Youngblut ND, Reischer GH, Walters W, Schuster N, Walzer C, Stalder G, Ley RE, Farnleitner AH (2019) Host diet and evolutionary history explain different aspects of gut microbiome diversity among vertebrate clades. Nat Commun 10:2200
McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ (2013) Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA 110:3229–3236
Ley RE, Hamady M, Lozupone CA, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI (2008) Evolution of mammals and their gut microbes. Science 320:1647–1651
Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, Henrissat B, Knight R, Gordon JI (2011) Diet convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332:970–974
Phillips CD, Phelan G, Dowd SE, McDonough MM, Ferguson AW, Hanson JD, Siles L, Ordóñez-Garza N, San Francisco M, Baker RJ (2012) Microbiome analysis among bats describes influences of host phylogeny, life history, physiology and geography. Mol Ecol 21:2617–2627
Shanahan T (2011) Phylogenetic inertia and Darwin’s higher law. Stud Hist PhilosSci 42:60–68
Carrillo-Araujo M, Tas N, Alcántara-Hernández RJ, Gaona O, Schondube JE, Medellín RA, Jansson JK, Falcón LI (2015) Phyllostomid bat microbiome composition is associated to host phylogeny and feeding strategies. Front Microbiol 6:447
Lutz HL, Jackson EW, Webala PW, Babyesiza WS, Kerbis Peterhans JC, Demos TC, Patterson BD, Gilbert JA (2019) Ecology and host identity outweigh evolutionary history in shaping the bat microbiome. mSystems 4:e00511-19
Ingala MR, Simmons NB, Wultsch C, Krampis K, Speer KA, Perkins SL (2018) Comparing microbiome sampling methods in a wild mammal: fecal and intestinal samples record different signals of host ecology, evolution. Front Microbiol 9:803
Devine DA, Marsh PD, Meade J (2015) Modulation of host responses by oral commensal bacteria. J Oral Microbiol 7:26941
Marsh PD (2018) In sickness and in health – What does the oral microbiome mean to us? an ecological perspective. AdvDent Res 29:60–65
Findley JS (1993) Bats: a community perspective. Cambridge University Press, Cambridge, UK
Galindo-González J, Guevara S, Sosa VJ (2000) Bat and bird-generated seed rains at isolated trees in pastures in a tropical rainforest. Conserv Biol 14:1693–1703
Cisneros LM, Fagan ME, Willig MR (2015) Effects of human-modified landscapes on taxonomic, functional, and phylogenetic dimensions of bat biodiversity. Divers Distrib 21:523–533
Presley SJ, Cisneros LM, Klingbeil BT, Willig MR (2019) Landscape ecology of mammals. J Mammal 100:1044–1068
Mühldorfer K (2013) Bats and bacterial pathogens: a review. Zoonoses Public Hlth 60:93–103
Olival KJ, Hosseini PR, Zambrana-Torrelio C, Ross N, Bogich TL, Daszak P (2017) Host and viral traits predict zoonotic spillover from mammals. Nature 546:646–650
Veikkolainen V, Vesterinen EJ, Lilley TM, Pulliainen AT (2014) Bats as reservoir hosts of human bacterial pathogen, Bartonella mayotimonensis. Emerg Infect Dis 20:960–967
Hahn MB, Gurley ES, Epstein JH, Islam MS, Patz JA, Daszak P, Luby SP (2014) The role of landscape composition and configuration on Pteropus giganteus roosting ecology and Nipah virus spillover risk in Bangladesh. Am J Trop Med Hyg 90:247–255
Fleming TH, Eby P (2003) Ecology of bat migration. In: Kunz TH, Fenton MB (eds) Bat ecology. University of Chicago Press, Chicago, pp 156–208
Caviedes-Vidal C, McWhorter TJ, Lavin SR, Chediack JG, Tracy CR, Karasov WH (2007) The digestive adaptation of flying vertebrates: high intestinal paracellular absorption compensates for smaller guts. Proc Natl Acad Sci USA 104:19132–19137
Gannon MR, Duran MR, Kurta A, Willig MR (2005) Bats of Puerto Rico: an Island Focus and Caribbean Perspective. Texas Tech University Press, Lubbock, TX
Teeling EC, Springer MS, Madsen O, Bates P, O’Brien SJ, Murphy WJ (2005) A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307:580–584
Jost L (2006) Entropy and diversity. Oikos 113:363–375
Vengust M, Knapic T, Weese JS (2018) The fecal bacterial microbiota of bats. Slovenia. PLoS One 13:e0196728
Hill MO (1973) Diversity and evenness: a unifying notation and its consequences. Ecology 54:427–432
Ma Z (2018) Measuring microbiome diversity and similarity with Hill numbers. In: Nagarajan M (ed) Metagenomics: perspectives, methods, and applications. Academic Press, Cambridge, MA, pp 157–178
Menzel MA, Carter TC, Jablonowski LR, Mitchell BL, Menzel JM, Chapman BR (2001) Home range size and habitat use of big brown bats (Eptesicus fuscus) in a maternity colony located on a rural-urban interface in the southeast. J Elisha Mitchell Sci Soc 117:36–45
Lewis SE (1995) Roost fidelity in bats: a review. J Mammal 76:481–496
Rodríguez-Duran A (2009) Bat assemblages in the West Indies: the role of caves. In: Fleming TH, Racey PA (eds) Island bats: evolution, ecology, and conservation. University of Chicago Press, Chicago, IL, pp 265–280
Benjamino J, Beka L, Graf J (2018) Microbiome analyses for toxicological studies. Curr Protoc Toxicol 77:e53
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108:4516–4522
R Core Team (2019) R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. URL https://www.R-project.org/
Schliep K, Paradis E, Martins LdO, Potts A, White TW, Stachniss C, Kendall M, Halabi K, Bilderbeek R, Winchell K (2019) phangorn: phylogenetic reconstruction and analysis. R Package version 2.5.5. http://cran.rproject.org/web/packages/phangorn
Davis NM, Proctor D, Holmes SP, Relman DA, Callahan BJ (2018) Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6:226
Magurran AE, McGill BJ (2011) Biological diversity. Oxford University Press, New York, NY
Kunz TH, August PV, Burnett CD (1983) Harem social organization in cave roosting Artibeus jamaicensis (Chiroptera: Phyllostomidae). Biotropica 15:133–138
Park KJ, Maters E, Altringham JD (1998) Social structure of three sympatric bat species (Vespertilionidae). J Zool 244:379–389
Lopez JE, Vaughan C (2007) Food niche overlap among Neotropical frugivorous bats in Costa Rica. Rev Biol Trop 55:301–313
López-González C, Presley SJ, Lozano A, Stevens RD, Higgins CL (2015) Ecological biogeography of Mexican bats: the relative contributions of habitat heterogeneity, beta diversity, and environmental gradients to species richness and composition patterns. Ecography 38:261–272
Igartua C, Davenport ER, Gilad Y, Nicolae DL, Pinto J, Ober C (2017) Host genetic variation in mucosal immunity pathways influences the upper airway microbiome. Microbiome 5:1–17
Goldford JE, Nanxi L, Bajić D, Estrela S, Tikhonov M, Sanchez-Grostiaga A, Segrè D, Mehta P, Sanchez A (2018) Emergent simplicity in microbial community assembly. Science 361:469–474
Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI (2008) Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol 6:776–788
Price ER, Brun A, Caviedes-Vidal E, Karasov WH (2015) Digestive adaptations of aerial lifestyles. Physiology 30:69–78
Speakman JR, Thomas DW (2003) Physiological ecology and energetics of bats. In: Kunz TH, Fenton MB (eds) Bat ecology. University of Chicago Press, Chicago, IL, pp 430–490
Herrera MLG, Martínez Del Río C (1998) Pollen digestion by new world bats: effects of processing time and feeding habits. Ecology 79:2828–2838
Schondube JE, Herrera MLG, Martínez del Río C (2001) Diet and the evolution of digestion and renal function in phyllostomid bats. Zoology 104:59–73
Caviedes-Vidal C, Karasov WH, Chediack JG, Fasulo V, Cruz-Neto AP, Otani L (2008) Paracellular absorption: a bat breaks the mammal paradigm. PloS One 1:e1425
Acknowledgements
We gratefully acknowledge our team of field volunteers, Armando Rodríguez-Duran for logistical help, and Para la Naturaleza for providing access to the Rio Encantado field site.
Funding
This work was supported by a Microbiome Research Seed Grant to MRW and JG from the Office of the Vice President of Research at the University of Connecticut. In addition, SJP and MRW were supported by the National Science Foundation (DEB-1546686 and DEB-1831952) as well as by the Center for Environmental Sciences and Engineering and Institute of the Environment at the University of Connecticut.
Author information
Authors and Affiliations
Contributions
MRW and JG conceived the project, and with SJP and ARS crafted the experimental design. ARS conducted sample collection. AFH conducted laboratory analyses. SJP conducted statistical analyses and wrote the manuscript. All authors critically reviewed multiple drafts of the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical Approval
All methods were approved by the University of Connecticut Institutional Animal Care and Use Committee (IACUC, protocol A15-032).
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Presley, S.J., Graf, J., Hassan, A.F. et al. Effects of Host Species Identity and Diet on the Biodiversity of the Microbiota in Puerto Rican Bats. Curr Microbiol 78, 3526–3540 (2021). https://doi.org/10.1007/s00284-021-02607-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-021-02607-5